Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
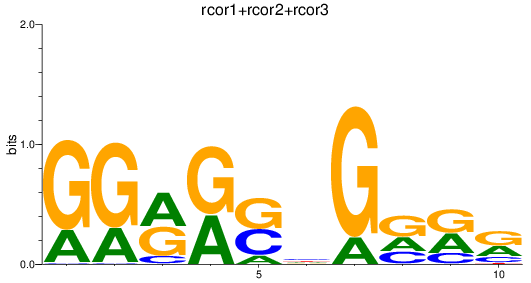
Results for rcor1+rcor2+rcor3
Z-value: 0.69
Transcription factors associated with rcor1+rcor2+rcor3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rcor3
|
ENSDARG00000004502 | REST corepressor 3 |
|
rcor2
|
ENSDARG00000008278 | REST corepressor 2 |
|
rcor1
|
ENSDARG00000031434 | REST corepressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rcor2 | dr11_v1_chr7_-_24994722_24994722 | 0.97 | 7.7e-03 | Click! |
| rcor3 | dr11_v1_chr22_+_38276024_38276024 | 0.96 | 1.0e-02 | Click! |
| rcor1 | dr11_v1_chr17_-_29271359_29271359 | 0.52 | 3.7e-01 | Click! |
Activity profile of rcor1+rcor2+rcor3 motif
Sorted Z-values of rcor1+rcor2+rcor3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_22535 | 2.02 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr11_-_44543082 | 0.88 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr16_-_27640995 | 0.81 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr16_-_17207754 | 0.55 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr4_+_21206283 | 0.55 |
ENSDART00000041861
|
syt1a
|
synaptotagmin Ia |
| chr15_-_12113045 | 0.53 |
ENSDART00000159879
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr17_-_22048233 | 0.51 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr6_+_58543336 | 0.49 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr19_-_27966780 | 0.49 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr6_-_58828113 | 0.49 |
ENSDART00000180934
|
kif5ab
|
kinesin family member 5A, b |
| chr11_-_44030962 | 0.48 |
ENSDART00000171910
|
FP016005.1
|
|
| chr2_-_44282796 | 0.47 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr24_+_32411753 | 0.46 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr20_+_66857 | 0.45 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr11_+_14676236 | 0.45 |
ENSDART00000109308
|
si:ch73-60h1.1
|
si:ch73-60h1.1 |
| chr23_+_44732863 | 0.45 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr8_+_16004154 | 0.42 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_58686860 | 0.41 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr25_+_21324588 | 0.40 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr19_-_27196090 | 0.38 |
ENSDART00000045616
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr1_-_39943596 | 0.38 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr7_+_528593 | 0.38 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr6_-_52156427 | 0.37 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr21_+_25643880 | 0.37 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr14_-_1949277 | 0.37 |
ENSDART00000159435
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr8_+_24861264 | 0.37 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr8_+_28065803 | 0.36 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr6_-_58828398 | 0.35 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr13_+_1100197 | 0.35 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr11_-_25734417 | 0.35 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr23_-_14990865 | 0.34 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr16_-_12914288 | 0.34 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr25_+_2993855 | 0.33 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr11_-_32723851 | 0.33 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr21_-_26918901 | 0.33 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr19_+_6938289 | 0.32 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr6_-_60031693 | 0.32 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr3_-_56933578 | 0.32 |
ENSDART00000192185
|
hid1a
|
HID1 domain containing a |
| chr3_+_22578369 | 0.32 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr22_+_110158 | 0.31 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr22_+_10215558 | 0.31 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr16_-_25285469 | 0.31 |
ENSDART00000183943
ENSDART00000191103 ENSDART00000154543 |
prelid3a
|
PRELI domain containing 3A |
| chr5_+_22098591 | 0.30 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr10_-_35542071 | 0.30 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr6_+_41255485 | 0.30 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr16_+_13822137 | 0.29 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr6_-_13408680 | 0.29 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr22_-_38508112 | 0.29 |
ENSDART00000192004
|
klc4
|
kinesin light chain 4 |
| chr22_+_12366516 | 0.28 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr19_+_41769135 | 0.28 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr3_+_33340939 | 0.28 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr5_-_32445835 | 0.27 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr11_+_11271959 | 0.27 |
ENSDART00000190678
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr8_+_29749017 | 0.27 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr8_+_21629941 | 0.27 |
ENSDART00000140145
|
ajap1
|
adherens junctions associated protein 1 |
| chr10_+_34377697 | 0.27 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr5_+_32009956 | 0.26 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr12_+_32159272 | 0.26 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr9_+_2393764 | 0.26 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr6_+_22597362 | 0.26 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr8_+_25267903 | 0.25 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr24_-_41320037 | 0.25 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr22_-_12160283 | 0.25 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr8_+_26868105 | 0.25 |
ENSDART00000005337
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr9_-_23824290 | 0.24 |
ENSDART00000059209
|
wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr24_-_26518972 | 0.24 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr9_-_27649406 | 0.24 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr19_+_9050852 | 0.24 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr2_+_50094873 | 0.23 |
ENSDART00000132307
|
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr10_-_1718395 | 0.23 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr16_-_24518027 | 0.23 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr21_+_43561650 | 0.23 |
ENSDART00000085071
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr14_-_1990290 | 0.22 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr14_-_2369849 | 0.22 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr20_+_13969414 | 0.22 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr19_+_14059349 | 0.22 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr12_+_32729470 | 0.22 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr21_-_14966718 | 0.21 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr6_-_21873266 | 0.21 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr9_+_21535885 | 0.21 |
ENSDART00000141408
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr19_+_9533008 | 0.21 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr11_+_36231248 | 0.21 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr7_+_21752168 | 0.21 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr7_+_39624728 | 0.21 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr4_+_9394426 | 0.21 |
ENSDART00000092013
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr20_-_1560724 | 0.21 |
ENSDART00000179193
ENSDART00000092508 ENSDART00000133369 |
ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr1_-_7894255 | 0.20 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr14_-_572742 | 0.20 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr17_-_38887424 | 0.20 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr7_-_38477235 | 0.20 |
ENSDART00000084355
|
zgc:165481
|
zgc:165481 |
| chr5_-_31772559 | 0.20 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr2_+_45374163 | 0.20 |
ENSDART00000193650
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr12_+_35203091 | 0.20 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr23_-_6722101 | 0.20 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr6_+_59991076 | 0.20 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr2_+_59015878 | 0.20 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr16_+_17389116 | 0.19 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr19_-_27776649 | 0.19 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr15_+_47418565 | 0.19 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr19_-_2029777 | 0.19 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr16_-_28658341 | 0.19 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr20_-_53321499 | 0.19 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr21_+_44581654 | 0.19 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr16_-_34258931 | 0.18 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr12_+_31494830 | 0.18 |
ENSDART00000153390
|
gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr10_+_21722892 | 0.18 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr7_+_36898622 | 0.18 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr7_+_56735195 | 0.18 |
ENSDART00000082830
|
KIAA0895L
|
KIAA0895 like |
| chr2_+_24867534 | 0.18 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr4_-_2868112 | 0.18 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr11_+_45299447 | 0.18 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr22_+_24157807 | 0.18 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr10_-_7347311 | 0.18 |
ENSDART00000168885
|
nrg1
|
neuregulin 1 |
| chr23_-_9965148 | 0.17 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr2_-_11258547 | 0.17 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr15_-_11956981 | 0.17 |
ENSDART00000164163
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr19_+_17400283 | 0.17 |
ENSDART00000127353
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr7_+_31051213 | 0.17 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr23_+_2825940 | 0.17 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr17_+_43629008 | 0.17 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr6_-_7726849 | 0.17 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr19_+_30867845 | 0.17 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr16_-_19890303 | 0.17 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr4_-_9728730 | 0.16 |
ENSDART00000150265
|
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr3_+_37827373 | 0.16 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr17_+_41992054 | 0.16 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr12_+_44610912 | 0.15 |
ENSDART00000179030
ENSDART00000178008 |
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr1_-_7893808 | 0.15 |
ENSDART00000110154
|
radil
|
Ras association and DIL domains |
| chr20_-_2355357 | 0.15 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr12_+_18971100 | 0.15 |
ENSDART00000164212
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr14_+_29550323 | 0.15 |
ENSDART00000157943
|
TENM2
|
si:dkey-34l15.2 |
| chr23_-_18017946 | 0.15 |
ENSDART00000104592
|
pm20d1.2
|
peptidase M20 domain containing 1, tandem duplicate 2 |
| chr3_+_9379877 | 0.15 |
ENSDART00000182080
|
LO018550.1
|
|
| chr25_-_32449235 | 0.15 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr14_-_2217285 | 0.15 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr10_-_5847904 | 0.14 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr14_+_34547554 | 0.14 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr12_+_13348918 | 0.14 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr14_+_24283915 | 0.14 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr2_+_11685742 | 0.14 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr10_-_35236949 | 0.14 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr7_-_33829824 | 0.14 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr19_+_7559901 | 0.14 |
ENSDART00000141189
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr3_+_60957512 | 0.14 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr6_-_42418225 | 0.14 |
ENSDART00000002501
|
ip6k2a
|
inositol hexakisphosphate kinase 2a |
| chr1_+_52929185 | 0.14 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr14_-_2004291 | 0.14 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr20_+_54333774 | 0.14 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr17_+_28624321 | 0.13 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr4_-_2014406 | 0.13 |
ENSDART00000180463
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr8_-_13177536 | 0.13 |
ENSDART00000100992
|
zgc:194990
|
zgc:194990 |
| chr12_+_14149686 | 0.13 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr13_+_31070181 | 0.13 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr4_+_12612145 | 0.13 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr23_+_2760573 | 0.13 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr19_+_49721 | 0.13 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr6_+_23809163 | 0.13 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr10_-_641609 | 0.13 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr22_-_11724563 | 0.12 |
ENSDART00000190796
ENSDART00000184744 |
krt222
|
keratin 222 |
| chr6_-_1820606 | 0.12 |
ENSDART00000183228
|
FO834857.1
|
|
| chr23_-_45504991 | 0.12 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr17_-_23412705 | 0.12 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr3_-_23596532 | 0.12 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr9_-_10532591 | 0.12 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr14_-_2196267 | 0.12 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr10_-_26179805 | 0.12 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr24_-_5910811 | 0.12 |
ENSDART00000163012
|
pcolce2b
|
procollagen C-endopeptidase enhancer 2b |
| chr15_-_33640890 | 0.12 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr15_-_33807758 | 0.12 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr23_+_22335407 | 0.12 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr18_-_44611252 | 0.12 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr13_-_18119696 | 0.12 |
ENSDART00000148125
|
washc2c
|
WASH complex subunit 2C |
| chr2_-_39759059 | 0.12 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr11_-_19694334 | 0.12 |
ENSDART00000054735
|
SYNPR
|
si:dkey-30j16.3 |
| chr5_+_51102010 | 0.12 |
ENSDART00000110377
|
zgc:194398
|
zgc:194398 |
| chr21_-_3201027 | 0.12 |
ENSDART00000161256
|
zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr6_+_3516695 | 0.11 |
ENSDART00000109576
|
faah
|
fatty acid amide hydrolase |
| chr7_+_60111581 | 0.11 |
ENSDART00000087093
|
hspa12b
|
heat shock protein 12B |
| chr7_-_35379899 | 0.11 |
ENSDART00000183484
|
slc6a2
|
solute carrier family 6 (neurotransmitter transporter), member 2 |
| chr2_-_7185460 | 0.11 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr25_+_20116407 | 0.11 |
ENSDART00000183615
ENSDART00000193243 |
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr20_-_26588736 | 0.11 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr12_-_7806007 | 0.11 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr3_-_23596809 | 0.11 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr24_-_26715174 | 0.11 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr10_+_40214877 | 0.10 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr4_-_23839789 | 0.10 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr13_-_4367083 | 0.10 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr18_+_17754510 | 0.10 |
ENSDART00000189372
ENSDART00000135236 ENSDART00000137239 |
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr14_+_52408619 | 0.10 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr22_-_876506 | 0.10 |
ENSDART00000137522
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr6_-_13709591 | 0.10 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr1_+_19535144 | 0.10 |
ENSDART00000103089
|
si:dkey-245p14.4
|
si:dkey-245p14.4 |
| chr12_+_47907277 | 0.10 |
ENSDART00000097714
|
tbata
|
thymus, brain and testes associated |
| chr15_+_31806303 | 0.10 |
ENSDART00000155902
|
frya
|
furry homolog a (Drosophila) |
| chr15_-_43768776 | 0.10 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr1_-_45584407 | 0.10 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr23_+_43770149 | 0.10 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr17_-_25630822 | 0.09 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr3_+_15776446 | 0.09 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rcor1+rcor2+rcor3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.8 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.3 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.3 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.4 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.3 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0005333 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |