Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
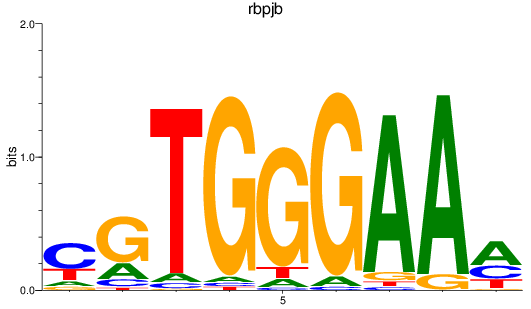
Results for rbpjb
Z-value: 1.50
Transcription factors associated with rbpjb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpjb
|
ENSDARG00000052091 | recombination signal binding protein for immunoglobulin kappa J region b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpjb | dr11_v1_chr7_+_62080456_62080456 | 0.28 | 6.5e-01 | Click! |
Activity profile of rbpjb motif
Sorted Z-values of rbpjb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_31891110 | 1.75 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr17_-_25326685 | 1.40 |
ENSDART00000082327
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr17_-_25326296 | 1.30 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr10_+_22782522 | 1.10 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr24_-_37877743 | 1.10 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr20_-_39596338 | 0.90 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr4_-_74367912 | 0.86 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr25_+_6122823 | 0.78 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr19_-_25113660 | 0.78 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr6_-_36552844 | 0.73 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr3_+_41917499 | 0.67 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr24_-_6375774 | 0.65 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr3_-_36839115 | 0.64 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr16_-_42523744 | 0.63 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr10_+_9195190 | 0.62 |
ENSDART00000136364
|
antxr2b
|
anthrax toxin receptor 2b |
| chr15_-_37846047 | 0.62 |
ENSDART00000184837
|
hsc70
|
heat shock cognate 70 |
| chr24_-_37877978 | 0.61 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr9_+_38684871 | 0.57 |
ENSDART00000147938
ENSDART00000004462 |
heg1
|
heart development protein with EGF-like domains 1 |
| chr4_-_9722568 | 0.56 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr24_-_37877554 | 0.55 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr20_+_36234335 | 0.55 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr22_+_16308450 | 0.53 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr6_+_45932276 | 0.51 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr11_+_41981959 | 0.51 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr18_+_22220656 | 0.50 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr5_+_23151169 | 0.50 |
ENSDART00000125638
|
tbx5b
|
T-box 5b |
| chr22_+_16308806 | 0.49 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr4_+_4849789 | 0.49 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr5_-_69312533 | 0.49 |
ENSDART00000082614
ENSDART00000183098 |
smtnb
|
smoothelin b |
| chr7_+_27834130 | 0.45 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr19_-_32641725 | 0.41 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr23_-_36003441 | 0.40 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr5_-_47727819 | 0.40 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr22_-_30881738 | 0.39 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr13_-_33009734 | 0.39 |
ENSDART00000134140
|
rbm25a
|
RNA binding motif protein 25a |
| chr19_+_42609132 | 0.37 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr13_-_36579086 | 0.36 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr2_+_6734108 | 0.33 |
ENSDART00000112227
|
brinp3b
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3b |
| chr14_-_33083539 | 0.33 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr23_+_31405497 | 0.32 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr13_+_30804367 | 0.31 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr16_+_36671064 | 0.30 |
ENSDART00000109703
|
col6a4a
|
collagen, type VI, alpha 4a |
| chr13_+_33246150 | 0.30 |
ENSDART00000136356
ENSDART00000132834 ENSDART00000144379 ENSDART00000131985 ENSDART00000148368 ENSDART00000035940 ENSDART00000141912 |
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr21_-_27213166 | 0.30 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr24_-_20658446 | 0.29 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr23_-_36003282 | 0.29 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr7_+_38445938 | 0.28 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr6_-_12172424 | 0.28 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr12_+_38563073 | 0.28 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
| chr1_-_45633955 | 0.27 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr2_+_1881334 | 0.25 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr2_-_44746723 | 0.25 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr2_-_898899 | 0.24 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr5_-_25072607 | 0.24 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr25_+_6186823 | 0.24 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr21_-_4849029 | 0.22 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr25_+_10793478 | 0.22 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr5_-_56513825 | 0.21 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr6_-_14292307 | 0.21 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr5_-_23675222 | 0.20 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr20_+_39283849 | 0.20 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr14_+_23076207 | 0.20 |
ENSDART00000161628
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr17_+_30546579 | 0.20 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr16_-_27628994 | 0.19 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr12_+_35587971 | 0.19 |
ENSDART00000166072
ENSDART00000181861 ENSDART00000158043 |
chmp6b
|
charged multivesicular body protein 6b |
| chr5_-_7834582 | 0.19 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr14_+_41406321 | 0.19 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr15_+_19783352 | 0.19 |
ENSDART00000178784
ENSDART00000092560 |
zpld1b
|
zona pellucida-like domain containing 1b |
| chr3_+_46315016 | 0.19 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr8_+_48491387 | 0.19 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr18_-_11184584 | 0.19 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr11_-_27962757 | 0.19 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr13_-_24874950 | 0.19 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr8_+_11993150 | 0.19 |
ENSDART00000132492
ENSDART00000191519 ENSDART00000114432 |
ntng2a
|
netrin g2a |
| chr22_+_30335936 | 0.18 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr8_-_29706882 | 0.18 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr7_-_26436436 | 0.18 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr23_-_23401305 | 0.18 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_+_34976600 | 0.17 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr25_+_3525494 | 0.17 |
ENSDART00000169775
|
RAB19 (1 of many)
|
RAB19, member RAS oncogene family |
| chr10_-_10863936 | 0.17 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr10_-_42297889 | 0.17 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr17_-_20287530 | 0.17 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr10_-_10864331 | 0.16 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr6_-_14139503 | 0.16 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr13_-_27916439 | 0.16 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr23_-_31372639 | 0.16 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr17_-_6730247 | 0.15 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr21_-_43485351 | 0.15 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr11_-_1409236 | 0.15 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr5_+_9428876 | 0.15 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr1_-_47071979 | 0.15 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr14_+_8475007 | 0.15 |
ENSDART00000148210
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr8_-_18613948 | 0.15 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr13_-_36798204 | 0.14 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr6_-_49861506 | 0.14 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr3_-_28665291 | 0.14 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr19_-_12648408 | 0.14 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr19_+_48018464 | 0.14 |
ENSDART00000172307
ENSDART00000163848 |
UBE2M
|
si:ch1073-205c8.3 |
| chr22_-_26834043 | 0.14 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr16_-_9869056 | 0.13 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr13_+_33655404 | 0.13 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr11_+_13159988 | 0.13 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr19_-_7540821 | 0.13 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr7_+_39634873 | 0.13 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr1_+_32528097 | 0.13 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr7_+_38770167 | 0.13 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr3_-_34547000 | 0.12 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr6_+_58522557 | 0.12 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr11_-_11518469 | 0.12 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr23_-_8733092 | 0.12 |
ENSDART00000014222
|
tcea2
|
transcription elongation factor A (SII), 2 |
| chr12_-_19862912 | 0.12 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr1_-_46924801 | 0.12 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr20_+_45620076 | 0.12 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr7_-_71837213 | 0.12 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr1_+_41886830 | 0.12 |
ENSDART00000185887
ENSDART00000084735 |
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr12_-_15584479 | 0.12 |
ENSDART00000150134
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr12_+_19362335 | 0.12 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr9_+_19623363 | 0.12 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr7_+_52712807 | 0.12 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr10_+_1668106 | 0.11 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr19_+_3842891 | 0.11 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr12_+_21299338 | 0.11 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr23_-_31346319 | 0.11 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr22_+_12431608 | 0.10 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr20_+_22799641 | 0.10 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr8_+_21159122 | 0.10 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr23_-_21471022 | 0.10 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr24_-_19718077 | 0.10 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr24_+_31374324 | 0.10 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr3_-_46379900 | 0.10 |
ENSDART00000006783
|
snrnp25
|
small nuclear ribonucleoprotein 25 |
| chr18_+_14277003 | 0.10 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr15_-_47468085 | 0.09 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr8_-_41273768 | 0.09 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr13_+_11440389 | 0.09 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_-_36597612 | 0.09 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr22_+_31930650 | 0.09 |
ENSDART00000092090
|
dock3
|
dedicator of cytokinesis 3 |
| chr24_+_28953089 | 0.09 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr22_-_16317886 | 0.09 |
ENSDART00000163664
|
trmt13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr11_-_7320211 | 0.09 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr16_-_27640995 | 0.09 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr20_+_6590220 | 0.09 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr14_+_4151379 | 0.09 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr8_-_41273461 | 0.09 |
ENSDART00000190598
|
rnf10
|
ring finger protein 10 |
| chr12_+_35067863 | 0.08 |
ENSDART00000152849
|
syt15
|
synaptotagmin XV |
| chr8_-_11067079 | 0.08 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr23_-_21446985 | 0.08 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr6_+_3680651 | 0.08 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr21_-_40562705 | 0.08 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr16_-_21047872 | 0.08 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr20_+_22799857 | 0.08 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr19_+_24896409 | 0.08 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr14_+_35806605 | 0.08 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr5_+_34578479 | 0.08 |
ENSDART00000016314
|
gfm2
|
G elongation factor, mitochondrial 2 |
| chr2_+_31957554 | 0.07 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr24_+_20658760 | 0.07 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr5_-_22082918 | 0.07 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr5_-_2752573 | 0.07 |
ENSDART00000168121
|
lzts3a
|
leucine zipper, putative tumor suppressor family member 3a |
| chr21_-_5799122 | 0.07 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr6_+_58522738 | 0.07 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr2_+_55916911 | 0.06 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr13_+_15657911 | 0.06 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr5_+_9348284 | 0.06 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr25_+_21178033 | 0.06 |
ENSDART00000041169
|
erc1a
|
ELKS/RAB6-interacting/CAST family member 1a |
| chr22_+_24715282 | 0.06 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr24_+_20658942 | 0.06 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr8_+_21225064 | 0.06 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr2_+_38608290 | 0.06 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr9_-_22355391 | 0.06 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr19_+_48018802 | 0.06 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr14_+_25817246 | 0.06 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr8_-_18899427 | 0.06 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr1_-_51720633 | 0.06 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr12_+_19362547 | 0.06 |
ENSDART00000180148
|
gspt1
|
G1 to S phase transition 1 |
| chr20_+_1349043 | 0.05 |
ENSDART00000186375
|
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr2_-_9646857 | 0.05 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr17_+_49081828 | 0.05 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr16_+_46492994 | 0.05 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr9_+_2020667 | 0.05 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr11_+_6159595 | 0.05 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr20_-_54508650 | 0.05 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr14_+_25817628 | 0.05 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr4_-_8779141 | 0.05 |
ENSDART00000182314
ENSDART00000172376 |
ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr9_-_3671911 | 0.05 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr24_+_33622769 | 0.04 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr24_-_24796583 | 0.04 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr25_-_20574859 | 0.04 |
ENSDART00000067288
|
adal
|
adenosine deaminase-like |
| chr13_-_33246154 | 0.04 |
ENSDART00000130127
|
rtf1
|
RTF1 homolog, Paf1/RNA polymerase II complex component |
| chr10_+_3507861 | 0.04 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr10_+_25982212 | 0.04 |
ENSDART00000128292
ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr3_-_41995321 | 0.04 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr5_+_44944778 | 0.04 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr5_+_33287611 | 0.03 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr1_+_10018466 | 0.03 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr5_+_61467618 | 0.03 |
ENSDART00000074073
ENSDART00000182094 |
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr20_-_15090862 | 0.03 |
ENSDART00000063892
ENSDART00000122592 |
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr19_+_40248697 | 0.03 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr11_+_34921492 | 0.03 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr7_+_53754653 | 0.03 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr3_+_21669545 | 0.03 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpjb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 0.9 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.2 | 0.5 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.6 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.2 | 0.8 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.9 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.2 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.2 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 1.0 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.1 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 0.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |