Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for rargb_pparab
Z-value: 2.31
Transcription factors associated with rargb_pparab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
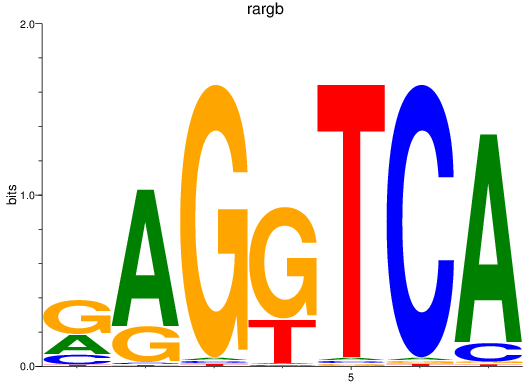
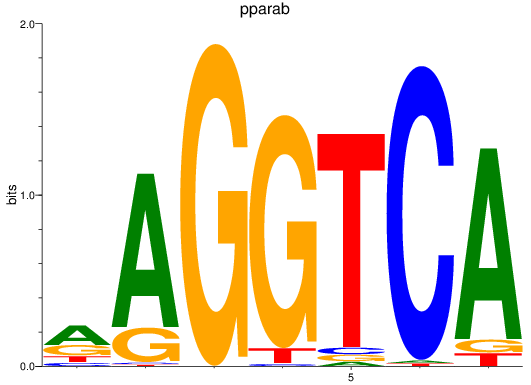
rargb
|
ENSDARG00000054003 | retinoic acid receptor, gamma b |
|
pparab
|
ENSDARG00000054323 | peroxisome proliferator-activated receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rargb | dr11_v1_chr11_+_2089461_2089461 | -0.45 | 4.4e-01 | Click! |
| pparab | dr11_v1_chr25_+_2229301_2229301 | -0.24 | 7.0e-01 | Click! |
Activity profile of rargb_pparab motif
Sorted Z-values of rargb_pparab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24289641 | 2.48 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr3_+_39568290 | 2.32 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr11_+_11201096 | 1.93 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr23_+_42810055 | 1.65 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr12_-_26415499 | 1.49 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr22_-_15593824 | 1.49 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr6_-_40722200 | 1.46 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr8_+_48613040 | 1.44 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr23_-_45705525 | 1.41 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr13_+_22480496 | 1.41 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_39566999 | 1.37 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_+_31871830 | 1.37 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr15_+_24644016 | 1.36 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr24_+_25467465 | 1.34 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr10_+_22782522 | 1.29 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr15_-_28618502 | 1.24 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr15_+_20403903 | 1.22 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr11_+_36243774 | 1.18 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr10_+_22775253 | 1.16 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr8_-_18535822 | 1.10 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr5_+_20693724 | 1.10 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr1_+_23783349 | 1.10 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr19_+_712127 | 1.08 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr13_-_37127970 | 1.05 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr22_-_15587360 | 0.99 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr15_-_20933574 | 0.96 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr11_+_25111846 | 0.96 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr23_-_35756115 | 0.92 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr13_+_22480857 | 0.90 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr11_+_24314148 | 0.90 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr18_-_226800 | 0.87 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr11_-_25213651 | 0.87 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr9_-_42861080 | 0.86 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr5_-_69316142 | 0.83 |
ENSDART00000157238
ENSDART00000144570 |
smtnb
|
smoothelin b |
| chr20_+_1996202 | 0.83 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr17_+_53424415 | 0.83 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr6_-_15653494 | 0.83 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr11_+_24313931 | 0.83 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr5_+_38276582 | 0.82 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr14_-_34771371 | 0.81 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr3_-_32590164 | 0.81 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr23_-_24488696 | 0.78 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr23_+_42813415 | 0.78 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr8_-_17980317 | 0.78 |
ENSDART00000129148
|
tnni3k
|
TNNI3 interacting kinase |
| chr10_+_6318227 | 0.77 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr17_-_5583345 | 0.77 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr2_-_44199722 | 0.76 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr24_-_28419444 | 0.76 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr6_-_1514767 | 0.75 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr11_-_7410537 | 0.73 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr4_+_2620751 | 0.73 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr22_+_10201826 | 0.72 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr6_-_40722480 | 0.72 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr7_+_60111581 | 0.71 |
ENSDART00000087093
|
hspa12b
|
heat shock protein 12B |
| chr6_-_49873020 | 0.71 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr12_-_20373058 | 0.71 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr8_-_979735 | 0.71 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr16_-_560574 | 0.70 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr18_+_36037223 | 0.70 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr15_+_24644251 | 0.69 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr3_+_24207243 | 0.69 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr23_+_18722915 | 0.69 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr8_-_17997845 | 0.69 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr1_-_55810730 | 0.68 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr11_+_42587900 | 0.67 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr21_-_18648861 | 0.67 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr25_+_25124684 | 0.66 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr20_+_19066596 | 0.66 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr5_-_46980651 | 0.66 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr13_+_281214 | 0.66 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr20_-_29864390 | 0.64 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr3_+_30257582 | 0.64 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr12_+_26670778 | 0.64 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr19_-_47555956 | 0.63 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr7_+_26716321 | 0.63 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr1_+_16144615 | 0.63 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr11_-_11471857 | 0.63 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr18_+_48423973 | 0.62 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr9_+_30464641 | 0.62 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr1_-_23110740 | 0.61 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr19_-_32487469 | 0.61 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr1_-_9109699 | 0.60 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr3_-_32170850 | 0.59 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr4_-_25836684 | 0.59 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr17_-_38778826 | 0.59 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr10_-_44017642 | 0.59 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr7_+_34297271 | 0.59 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr24_+_16393302 | 0.58 |
ENSDART00000188670
ENSDART00000081759 ENSDART00000177790 |
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_-_37681824 | 0.58 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr13_-_39947335 | 0.58 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr4_+_38344 | 0.58 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr20_-_29483514 | 0.57 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr17_+_23554932 | 0.57 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr20_+_54738210 | 0.57 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr18_-_16791331 | 0.56 |
ENSDART00000148222
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr24_+_792429 | 0.55 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr6_-_49898881 | 0.55 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr5_-_18007077 | 0.55 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr19_-_32783373 | 0.55 |
ENSDART00000145790
|
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr10_+_23060391 | 0.55 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr11_+_7158723 | 0.55 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr6_+_8176486 | 0.54 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr18_+_48608366 | 0.54 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr7_+_34296789 | 0.54 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr9_-_15424639 | 0.53 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr6_-_39764995 | 0.53 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr20_+_19066858 | 0.52 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr5_-_30475011 | 0.52 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr20_+_40150612 | 0.52 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr16_-_27138478 | 0.51 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr25_-_13363286 | 0.51 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr1_+_36612660 | 0.51 |
ENSDART00000190784
|
ednraa
|
endothelin receptor type Aa |
| chr18_+_27926839 | 0.51 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr15_-_29388012 | 0.51 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr6_+_52804267 | 0.50 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr14_-_29799993 | 0.50 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr12_-_4070058 | 0.50 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr23_+_18722715 | 0.50 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr19_-_37154436 | 0.50 |
ENSDART00000103155
|
cx39.4
|
connexin 39.4 |
| chr19_+_41169996 | 0.50 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr14_+_30340251 | 0.49 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr21_+_15704556 | 0.49 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr11_-_6970107 | 0.49 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr12_-_34827477 | 0.48 |
ENSDART00000153026
|
NDUFAF8
|
si:dkey-21c1.6 |
| chr13_+_28821841 | 0.48 |
ENSDART00000179900
|
CU639469.1
|
|
| chr14_+_33413980 | 0.48 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr8_-_14184423 | 0.48 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr2_-_48753873 | 0.48 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr4_-_7212875 | 0.47 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr6_+_41463786 | 0.47 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr1_-_17650223 | 0.47 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr6_-_10912424 | 0.46 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr14_-_33454595 | 0.46 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr16_-_17197546 | 0.46 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr13_+_50375800 | 0.45 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr2_+_38039857 | 0.45 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr4_-_5597167 | 0.45 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr3_+_20001608 | 0.45 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr1_+_13930625 | 0.45 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr4_-_20235904 | 0.45 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr17_-_31058664 | 0.44 |
ENSDART00000126414
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr22_-_834106 | 0.44 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr6_-_39765546 | 0.44 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_25913225 | 0.44 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr8_+_31821396 | 0.44 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr16_-_27149202 | 0.44 |
ENSDART00000179726
|
tmem245
|
transmembrane protein 245 |
| chr3_+_14463941 | 0.44 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr19_+_2275019 | 0.43 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr7_+_5494565 | 0.43 |
ENSDART00000135271
|
MYADM
|
si:dkeyp-67a8.2 |
| chr11_+_77526 | 0.43 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr25_+_29160102 | 0.43 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr21_+_5080789 | 0.43 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr3_-_18410968 | 0.42 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr9_-_23944470 | 0.42 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr6_-_4214297 | 0.42 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr6_+_202367 | 0.42 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr4_-_4932619 | 0.42 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr1_-_11913170 | 0.41 |
ENSDART00000165543
|
grk4
|
G protein-coupled receptor kinase 4 |
| chr17_+_11675362 | 0.41 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr4_-_67799941 | 0.41 |
ENSDART00000185830
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr6_+_48618512 | 0.41 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr4_-_8152746 | 0.41 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr25_-_18953322 | 0.41 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr3_-_16010968 | 0.41 |
ENSDART00000080672
|
mrps34
|
mitochondrial ribosomal protein S34 |
| chr8_+_50534948 | 0.40 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr21_-_23035120 | 0.40 |
ENSDART00000016502
|
dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr15_+_404891 | 0.40 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr18_+_40364732 | 0.40 |
ENSDART00000123661
ENSDART00000130847 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr3_+_27770110 | 0.39 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr10_-_39153959 | 0.39 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr5_-_23280098 | 0.39 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr10_+_26834985 | 0.39 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr18_+_17959681 | 0.39 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr24_-_38374744 | 0.39 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr16_+_29650698 | 0.39 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr18_-_44526940 | 0.38 |
ENSDART00000077125
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr13_-_24880525 | 0.38 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr25_+_25766033 | 0.38 |
ENSDART00000103638
ENSDART00000039952 |
idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr18_+_30507839 | 0.38 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr23_+_5524247 | 0.38 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr20_-_39103119 | 0.38 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr21_-_20381481 | 0.38 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr5_+_21211135 | 0.38 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr9_-_710896 | 0.38 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr3_-_12187245 | 0.37 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr23_-_15216654 | 0.37 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr10_+_45128375 | 0.37 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr9_+_24106469 | 0.37 |
ENSDART00000039295
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr8_-_18010097 | 0.37 |
ENSDART00000122730
ENSDART00000133666 |
acot11b
|
acyl-CoA thioesterase 11b |
| chr18_-_44527124 | 0.37 |
ENSDART00000189471
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr23_-_35069805 | 0.37 |
ENSDART00000087219
|
BX294434.1
|
|
| chr24_+_35183595 | 0.36 |
ENSDART00000075142
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr14_+_1014109 | 0.36 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr12_+_26632448 | 0.36 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr18_+_25752592 | 0.36 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr20_+_23408970 | 0.36 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr25_-_31396479 | 0.36 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr23_+_17417539 | 0.36 |
ENSDART00000182605
|
BX649300.2
|
|
| chr14_-_17121676 | 0.36 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr22_-_26595027 | 0.36 |
ENSDART00000184162
|
CABZ01072309.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of rargb_pparab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.4 | 1.2 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.4 | 1.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 1.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.2 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.3 | 1.9 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.3 | 1.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 0.8 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.2 | 1.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 5.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 0.7 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 1.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 1.2 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.2 | 0.6 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.5 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 4.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.3 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.4 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.5 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.4 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.2 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 2.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.1 | GO:1905067 | negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.4 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 0.5 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.5 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.6 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 0.3 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.2 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.3 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.2 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.2 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.2 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 1.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.2 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.2 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.3 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.2 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:2000329 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.9 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 2.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.9 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.6 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0007620 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.6 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 1.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.7 | GO:0099587 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.0 | 0.1 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.7 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.3 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.0 | 0.1 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 3.2 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 2.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.4 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.7 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.8 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.8 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.1 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) positive regulation of cation transmembrane transport(GO:1904064) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 0.9 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.3 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 4.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.7 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 4.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.1 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.1 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.6 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 4.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 1.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 1.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 6.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.5 | GO:0030427 | site of polarized growth(GO:0030427) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.2 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 0.9 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.3 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.2 | 0.6 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.5 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.2 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 1.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.7 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 2.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.5 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.1 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 2.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:1990931 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.7 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.9 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 12.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 1.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 6.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 0.3 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.8 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0015491 | solute:cation antiporter activity(GO:0015298) cation:cation antiporter activity(GO:0015491) |
| 0.0 | 0.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |