Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
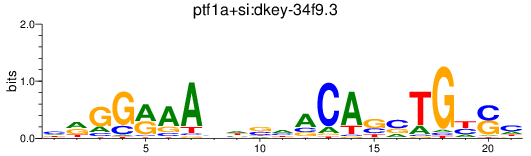
Results for ptf1a+si:dkey-34f9.3
Z-value: 1.97
Transcription factors associated with ptf1a+si:dkey-34f9.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ptf1a
|
ENSDARG00000014479 | pancreas associated transcription factor 1a |
|
si_dkey-34f9.3
|
ENSDARG00000077813 | si_dkey-34f9.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-34f9.3 | dr11_v1_chr9_+_45789887_45789887 | -0.87 | 5.7e-02 | Click! |
| ptf1a | dr11_v1_chr2_+_29710857_29710857 | -0.87 | 5.7e-02 | Click! |
Activity profile of ptf1a+si:dkey-34f9.3 motif
Sorted Z-values of ptf1a+si:dkey-34f9.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_5490668 | 2.46 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr7_-_38644560 | 2.36 |
ENSDART00000114934
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr7_-_38644287 | 2.27 |
ENSDART00000182307
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr22_-_20695237 | 1.49 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr24_+_6353394 | 1.42 |
ENSDART00000165118
|
CR352329.1
|
|
| chr13_+_33452096 | 1.35 |
ENSDART00000057370
|
ftr80
|
finTRIM family, member 80 |
| chr16_+_35905031 | 1.34 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr10_-_45029041 | 1.31 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr3_-_4579214 | 1.29 |
ENSDART00000164456
|
ftr50
|
finTRIM family, member 50 |
| chr2_-_40199780 | 1.24 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr9_-_17417628 | 1.18 |
ENSDART00000060425
ENSDART00000141997 |
ftr53
|
finTRIM family, member 53 |
| chr20_+_7084154 | 1.17 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr11_-_27962757 | 1.17 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr24_-_6029314 | 1.14 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr25_+_31227747 | 1.10 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr7_+_21841037 | 1.10 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr4_+_72797711 | 1.07 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr10_-_15048781 | 1.06 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr3_-_4591643 | 1.05 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr4_-_76488581 | 1.04 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr22_-_17489040 | 1.03 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr13_+_9100 | 1.00 |
ENSDART00000165772
|
ppp4r3b
|
protein phosphatase 4, regulatory subunit 3B |
| chr25_-_37314700 | 1.00 |
ENSDART00000017807
|
igl4v8
|
immunoglobulin light 4 variable 8 |
| chr2_-_43151358 | 0.99 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr2_+_47935476 | 0.97 |
ENSDART00000140555
|
ftr26
|
finTRIM family, member 26 |
| chr7_-_38638809 | 0.96 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr8_+_3379815 | 0.96 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr12_-_6172154 | 0.95 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr2_-_59231017 | 0.95 |
ENSDART00000143980
ENSDART00000184190 |
ftr29
|
finTRIM family, member 29 |
| chr2_-_59345920 | 0.94 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr12_-_9132682 | 0.91 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr19_+_7895086 | 0.88 |
ENSDART00000132180
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr3_-_54524194 | 0.84 |
ENSDART00000155406
ENSDART00000111791 |
si:ch73-208g10.1
|
si:ch73-208g10.1 |
| chr3_-_48980319 | 0.84 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr22_+_22438783 | 0.82 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr3_+_301479 | 0.79 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr15_-_35134918 | 0.78 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr4_-_58964138 | 0.78 |
ENSDART00000150259
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr15_+_12435975 | 0.76 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr18_-_1414760 | 0.76 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr23_-_6865946 | 0.75 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr3_+_3545825 | 0.73 |
ENSDART00000109060
|
CR589947.1
|
|
| chr25_+_37297659 | 0.73 |
ENSDART00000086733
|
si:dkey-234i14.13
|
si:dkey-234i14.13 |
| chr1_-_58009216 | 0.70 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr1_-_6085750 | 0.70 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr4_-_38281909 | 0.69 |
ENSDART00000162448
|
si:ch211-229l10.9
|
si:ch211-229l10.9 |
| chr3_-_4557653 | 0.69 |
ENSDART00000111305
|
ftr50
|
finTRIM family, member 50 |
| chr15_+_12436220 | 0.68 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr7_+_49664174 | 0.65 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_5904020 | 0.65 |
ENSDART00000172847
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr4_+_38981587 | 0.65 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr8_-_36554675 | 0.64 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr21_-_1642739 | 0.64 |
ENSDART00000151118
|
zgc:152948
|
zgc:152948 |
| chr5_+_28849155 | 0.63 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_+_28857969 | 0.62 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr2_+_42236118 | 0.60 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr5_+_28848870 | 0.60 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr2_-_47904043 | 0.60 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr2_+_42260021 | 0.59 |
ENSDART00000124702
ENSDART00000140203 ENSDART00000184079 ENSDART00000193349 |
ftr04
|
finTRIM family, member 4 |
| chr19_-_12212692 | 0.59 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr11_-_2478374 | 0.58 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr2_-_59265521 | 0.57 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr16_-_16152199 | 0.57 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr2_-_59327299 | 0.57 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr23_+_5736226 | 0.56 |
ENSDART00000134527
ENSDART00000112220 |
ftr57
|
finTRIM family, member 57 |
| chr5_+_37854685 | 0.56 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr21_-_20733615 | 0.54 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr9_+_3153717 | 0.54 |
ENSDART00000191958
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr16_-_46572834 | 0.54 |
ENSDART00000115248
|
zgc:174938
|
zgc:174938 |
| chr2_-_43130812 | 0.54 |
ENSDART00000039079
ENSDART00000056162 |
ftr13
|
finTRIM family, member 13 |
| chr2_-_42628028 | 0.53 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr22_-_9677264 | 0.52 |
ENSDART00000181737
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr2_-_14793343 | 0.52 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr16_-_46573211 | 0.52 |
ENSDART00000193392
|
zgc:174938
|
zgc:174938 |
| chr2_-_59205393 | 0.52 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr4_-_65323874 | 0.51 |
ENSDART00000175271
ENSDART00000187913 |
BX571813.1
|
|
| chr2_+_47927026 | 0.51 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr11_+_45347961 | 0.51 |
ENSDART00000172962
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr17_+_21477892 | 0.49 |
ENSDART00000155309
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr9_+_343062 | 0.49 |
ENSDART00000164012
ENSDART00000164365 |
bpifcl
|
BPI fold containing family C, like |
| chr16_-_46567344 | 0.48 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr7_+_24523017 | 0.48 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr9_+_3153466 | 0.48 |
ENSDART00000135619
|
si:zfos-979f1.2
|
si:zfos-979f1.2 |
| chr1_-_59287410 | 0.48 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr7_+_44608478 | 0.47 |
ENSDART00000149981
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr7_+_41812190 | 0.47 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr2_+_42294944 | 0.47 |
ENSDART00000140599
ENSDART00000178811 |
ftr06
|
finTRIM family, member 6 |
| chr18_-_45736 | 0.47 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr12_+_18663154 | 0.46 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr4_-_7231480 | 0.46 |
ENSDART00000159436
|
si:ch211-170p16.1
|
si:ch211-170p16.1 |
| chr6_-_58975010 | 0.45 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr2_-_59376399 | 0.45 |
ENSDART00000137134
|
ftr38
|
finTRIM family, member 38 |
| chr4_+_37406676 | 0.45 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr16_-_11961325 | 0.44 |
ENSDART00000143845
|
si:ch73-296e2.3
|
si:ch73-296e2.3 |
| chr5_+_28858345 | 0.44 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr18_-_18543358 | 0.43 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr12_-_3940768 | 0.43 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr7_+_41812817 | 0.40 |
ENSDART00000174165
|
orc6
|
origin recognition complex, subunit 6 |
| chr18_+_3140682 | 0.40 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr18_-_502722 | 0.38 |
ENSDART00000185757
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr22_-_36690742 | 0.37 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr21_+_20356458 | 0.36 |
ENSDART00000135816
|
pde6b
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr4_-_33525219 | 0.36 |
ENSDART00000150491
|
CT027801.2
|
|
| chr7_+_29177191 | 0.36 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr7_+_41812636 | 0.35 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr5_+_62723233 | 0.34 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr8_+_14959587 | 0.33 |
ENSDART00000138548
ENSDART00000041697 |
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr15_-_37610434 | 0.32 |
ENSDART00000156856
|
CR848763.1
|
|
| chr25_+_37285737 | 0.29 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr23_+_4890693 | 0.29 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr2_-_59178742 | 0.29 |
ENSDART00000170594
ENSDART00000109246 |
ftr95
|
finTRIM family, member 95 |
| chr15_-_41523225 | 0.29 |
ENSDART00000141202
ENSDART00000099290 ENSDART00000170395 ENSDART00000099437 |
si:dkey-230i18.2
si:dkey-121n8.7
nlrc9
|
si:dkey-230i18.2 si:dkey-121n8.7 NLR family CARD domain containing 9 |
| chr3_-_34624745 | 0.28 |
ENSDART00000151091
|
tac4
|
tachykinin 4 (hemokinin) |
| chr15_-_34933560 | 0.28 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr18_-_48530221 | 0.27 |
ENSDART00000188134
ENSDART00000142107 |
kcnj1a.2
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 2 |
| chr25_+_36336920 | 0.26 |
ENSDART00000073402
|
hist1h2a1
|
histone cluster 1 H2A family member1 |
| chr13_-_25196758 | 0.26 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr7_+_44608224 | 0.24 |
ENSDART00000005033
|
cmtm3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr3_+_37649436 | 0.24 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr3_+_5504990 | 0.23 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr4_+_69801372 | 0.22 |
ENSDART00000154125
|
si:dkey-238o14.7
|
si:dkey-238o14.7 |
| chr21_+_45819662 | 0.22 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr15_-_37620167 | 0.21 |
ENSDART00000059582
|
si:ch211-137j23.8
|
si:ch211-137j23.8 |
| chr25_-_15496485 | 0.21 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr22_+_15633013 | 0.21 |
ENSDART00000188095
ENSDART00000048763 |
CABZ01090041.1
|
|
| chr4_+_77681389 | 0.20 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr4_+_40018440 | 0.20 |
ENSDART00000110827
|
si:dkey-199k11.6
|
si:dkey-199k11.6 |
| chr3_+_61522759 | 0.19 |
ENSDART00000106266
|
znf1004
|
zinc finger protein 1004 |
| chr18_-_16801033 | 0.19 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr12_-_37343858 | 0.19 |
ENSDART00000153281
|
tekt3
|
tektin 3 |
| chr15_-_5212930 | 0.18 |
ENSDART00000136845
|
or128-6
|
odorant receptor, family E, subfamily 128, member 6 |
| chr8_+_4333914 | 0.18 |
ENSDART00000137528
|
myl2b
|
myosin, light chain 2b, regulatory, cardiac, slow |
| chr3_+_62196672 | 0.18 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr1_+_57311901 | 0.18 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr3_-_20721064 | 0.18 |
ENSDART00000193532
|
spop
|
speckle-type POZ protein |
| chr1_+_26467071 | 0.17 |
ENSDART00000112329
ENSDART00000159318 ENSDART00000193833 ENSDART00000011809 |
uso1
|
USO1 vesicle transport factor |
| chr25_-_34670413 | 0.16 |
ENSDART00000073440
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr16_-_30932533 | 0.16 |
ENSDART00000085761
|
slc45a4
|
solute carrier family 45, member 4 |
| chr2_-_1514001 | 0.16 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr15_+_31911989 | 0.15 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr17_-_86548 | 0.15 |
ENSDART00000163657
|
grem1b
|
gremlin 1b |
| chr8_+_16758304 | 0.14 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr4_-_76488854 | 0.12 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr24_+_26088143 | 0.12 |
ENSDART00000171129
|
CR352209.1
|
|
| chr15_-_2519640 | 0.11 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr11_-_27827442 | 0.11 |
ENSDART00000121847
ENSDART00000132018 ENSDART00000145744 ENSDART00000134677 ENSDART00000130800 |
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr2_+_11670270 | 0.10 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr24_+_34085940 | 0.10 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr4_-_41772315 | 0.09 |
ENSDART00000193109
|
znf976
|
zinc finger protein 976 |
| chr22_+_5123479 | 0.09 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr19_-_617246 | 0.08 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr3_+_57788761 | 0.07 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr11_-_16021424 | 0.07 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr3_-_58585854 | 0.07 |
ENSDART00000123546
|
CABZ01038521.1
|
|
| chr15_+_2475894 | 0.07 |
ENSDART00000035939
|
panx1a
|
pannexin 1a |
| chr23_-_28692581 | 0.06 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr15_+_33851280 | 0.04 |
ENSDART00000162308
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr10_+_10386435 | 0.03 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr15_-_46828958 | 0.03 |
ENSDART00000045110
|
chrd
|
chordin |
| chr14_-_30390145 | 0.01 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ptf1a+si:dkey-34f9.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.5 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.6 | GO:2000252 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.9 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.5 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 2.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.4 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 1.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.6 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 1.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 1.1 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 6.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 6.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 17.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |