Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
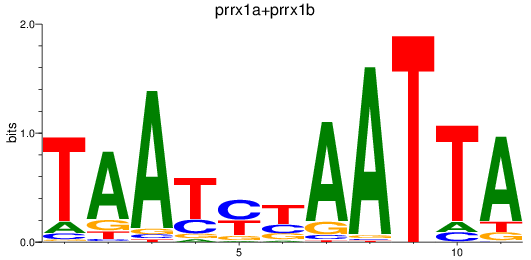
Results for prrx1a+prrx1b
Z-value: 0.86
Transcription factors associated with prrx1a+prrx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prrx1a
|
ENSDARG00000033971 | paired related homeobox 1a |
|
prrx1b
|
ENSDARG00000042027 | paired related homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prrx1b | dr11_v1_chr20_+_34512130_34512130 | -0.66 | 2.3e-01 | Click! |
| prrx1a | dr11_v1_chr2_-_23172708_23172708 | -0.53 | 3.6e-01 | Click! |
Activity profile of prrx1a+prrx1b motif
Sorted Z-values of prrx1a+prrx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 1.82 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_+_17676745 | 0.63 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr16_-_22294265 | 0.60 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr6_+_23887314 | 0.56 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr2_+_38039857 | 0.50 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr20_+_31287356 | 0.49 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr25_+_31267268 | 0.47 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr11_+_36243774 | 0.47 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr23_+_26026383 | 0.45 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr21_+_27382893 | 0.43 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr21_+_6556635 | 0.38 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr7_+_66822229 | 0.37 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr2_-_39558643 | 0.36 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr18_+_5490668 | 0.36 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr2_+_30916188 | 0.35 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr11_-_34097418 | 0.34 |
ENSDART00000028301
ENSDART00000138579 |
tdo2b
|
tryptophan 2,3-dioxygenase b |
| chr10_+_23537576 | 0.34 |
ENSDART00000130515
|
CR847844.1
|
|
| chr13_-_31622195 | 0.33 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr16_-_42872571 | 0.33 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr25_+_29160102 | 0.32 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr2_+_16781015 | 0.32 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr1_-_56080112 | 0.31 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr25_+_31277415 | 0.30 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr16_+_23984755 | 0.30 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr22_-_23668356 | 0.29 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr2_+_37227011 | 0.29 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr6_+_6924637 | 0.29 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr21_-_20757866 | 0.28 |
ENSDART00000163003
|
ghrb
|
growth hormone receptor b |
| chr7_+_20147202 | 0.28 |
ENSDART00000011398
|
si:ch73-335l21.1
|
si:ch73-335l21.1 |
| chr13_+_24279021 | 0.27 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr16_-_27749172 | 0.27 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr15_-_21165237 | 0.26 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr15_-_14625373 | 0.25 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr14_-_21123551 | 0.25 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr19_-_5669122 | 0.25 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr17_+_30704068 | 0.25 |
ENSDART00000062793
|
apoba
|
apolipoprotein Ba |
| chr22_+_21324398 | 0.25 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr12_-_25294769 | 0.24 |
ENSDART00000153306
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr7_+_41303572 | 0.24 |
ENSDART00000012692
|
rgs9bp
|
regulator of G protein signaling 9 binding protein |
| chr11_+_13224281 | 0.24 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr2_+_20605186 | 0.23 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr17_+_43863708 | 0.23 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr23_+_41679586 | 0.23 |
ENSDART00000067662
|
CU914487.1
|
|
| chr8_-_39739056 | 0.23 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr6_+_43234213 | 0.23 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr1_-_49250490 | 0.22 |
ENSDART00000150386
|
si:ch73-6k14.2
|
si:ch73-6k14.2 |
| chr9_-_394088 | 0.21 |
ENSDART00000169014
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr23_+_28770225 | 0.21 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr7_+_56098590 | 0.21 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr24_+_21346796 | 0.21 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr2_+_20604775 | 0.20 |
ENSDART00000131501
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr11_+_2391649 | 0.20 |
ENSDART00000104571
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr4_-_2945306 | 0.20 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr25_-_13728111 | 0.20 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr8_+_50190742 | 0.19 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr8_+_15254564 | 0.19 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr12_+_2428247 | 0.19 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr25_+_34641536 | 0.19 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr2_-_38284648 | 0.18 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr22_-_910926 | 0.18 |
ENSDART00000180075
|
FP016205.1
|
|
| chr4_+_2267641 | 0.18 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr19_-_25113660 | 0.18 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr21_-_42831033 | 0.18 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr10_+_34001444 | 0.18 |
ENSDART00000149934
|
kl
|
klotho |
| chr24_-_38657683 | 0.18 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr24_-_25098719 | 0.18 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr16_+_29509133 | 0.18 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr16_-_42066523 | 0.18 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr1_+_12009673 | 0.18 |
ENSDART00000080100
|
slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr23_+_39695827 | 0.18 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr5_+_57924611 | 0.18 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr6_+_41191482 | 0.17 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr11_+_13223625 | 0.17 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr23_+_45282858 | 0.17 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr22_-_24284447 | 0.17 |
ENSDART00000149894
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr6_+_8626427 | 0.17 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr10_-_21545091 | 0.16 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr22_-_38274188 | 0.16 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr14_-_46173265 | 0.16 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr25_-_10769039 | 0.15 |
ENSDART00000186758
ENSDART00000121724 |
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr1_-_18811517 | 0.15 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr5_+_20613466 | 0.15 |
ENSDART00000161209
ENSDART00000136632 |
CMKLR1 (1 of many)
|
si:ch211-240b21.5 |
| chr9_+_29548195 | 0.15 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr18_-_8030073 | 0.15 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr23_-_36441693 | 0.15 |
ENSDART00000024354
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr10_+_45031398 | 0.15 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr1_-_9195629 | 0.15 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr25_+_20089986 | 0.15 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr16_-_16761164 | 0.15 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr25_+_3327071 | 0.15 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr19_-_47323267 | 0.15 |
ENSDART00000190077
|
CU138512.1
|
|
| chr12_-_25294096 | 0.15 |
ENSDART00000183398
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr16_-_31598771 | 0.15 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr8_+_25034544 | 0.15 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr5_-_43859148 | 0.14 |
ENSDART00000162746
ENSDART00000128763 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr20_+_37393134 | 0.14 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr6_+_37894220 | 0.14 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr5_-_7199998 | 0.14 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr8_-_18203274 | 0.14 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr2_-_51303937 | 0.14 |
ENSDART00000164111
|
si:ch211-215e19.8
|
si:ch211-215e19.8 |
| chr4_-_77331797 | 0.14 |
ENSDART00000162325
|
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr3_+_32594357 | 0.13 |
ENSDART00000151639
|
unc119.2
|
unc-119 lipid binding chaperone B homolog 2 |
| chr21_-_28439596 | 0.13 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_-_38274995 | 0.13 |
ENSDART00000179786
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr25_+_31227747 | 0.13 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr21_-_20733615 | 0.13 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr6_+_3334710 | 0.13 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr3_-_32170850 | 0.13 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr9_+_47966167 | 0.13 |
ENSDART00000183345
|
gpbar1
|
G protein-coupled bile acid receptor 1 |
| chr20_+_23498255 | 0.13 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr12_+_27231607 | 0.13 |
ENSDART00000066270
|
tmem106a
|
transmembrane protein 106A |
| chr21_+_5272059 | 0.13 |
ENSDART00000141981
|
loxhd1a
|
lipoxygenase homology domains 1a |
| chr25_+_10923100 | 0.13 |
ENSDART00000157055
|
si:ch211-147g22.7
|
si:ch211-147g22.7 |
| chr7_-_55648336 | 0.12 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr5_+_40224938 | 0.12 |
ENSDART00000142897
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr13_-_9091354 | 0.12 |
ENSDART00000140319
ENSDART00000142697 |
HTRA2 (1 of many)
|
si:dkey-112g5.13 |
| chr15_+_17030941 | 0.12 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr14_-_29826659 | 0.12 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr24_-_25442342 | 0.12 |
ENSDART00000177140
ENSDART00000138215 |
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr11_-_26375575 | 0.12 |
ENSDART00000079255
|
utp3
|
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr15_-_18213515 | 0.12 |
ENSDART00000101635
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr5_-_57655092 | 0.12 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr3_-_16493528 | 0.12 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr21_+_26720803 | 0.12 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr1_-_43684584 | 0.12 |
ENSDART00000074609
|
suclg1
|
succinate-CoA ligase, alpha subunit |
| chr20_-_30920356 | 0.12 |
ENSDART00000022951
|
kif25
|
kinesin family member 25 |
| chr17_-_13058515 | 0.12 |
ENSDART00000172450
ENSDART00000170255 |
CU469462.1
|
|
| chr10_-_34107919 | 0.11 |
ENSDART00000167428
ENSDART00000145279 |
pimr144
pimr146
|
Pim proto-oncogene, serine/threonine kinase, related 144 Pim proto-oncogene, serine/threonine kinase, related 146 |
| chr12_+_27232173 | 0.11 |
ENSDART00000193714
|
tmem106a
|
transmembrane protein 106A |
| chr14_+_5835134 | 0.11 |
ENSDART00000054867
|
aup1
|
ancient ubiquitous protein 1 |
| chr3_+_16922226 | 0.11 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr1_+_15204633 | 0.11 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr1_-_45049603 | 0.11 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr23_+_32101202 | 0.11 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr9_+_55455801 | 0.11 |
ENSDART00000144757
ENSDART00000186543 |
mxra5b
|
matrix-remodelling associated 5b |
| chr24_+_24086491 | 0.11 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr22_+_1821718 | 0.11 |
ENSDART00000132220
|
znf1002
|
zinc finger protein 1002 |
| chr15_+_36096689 | 0.11 |
ENSDART00000049849
|
col4a3
|
collagen, type IV, alpha 3 |
| chr22_+_5971404 | 0.11 |
ENSDART00000122153
|
BX322587.1
|
|
| chr5_-_39698224 | 0.11 |
ENSDART00000076929
|
prkg2
|
protein kinase, cGMP-dependent, type II |
| chr23_-_31763753 | 0.11 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr3_+_26019426 | 0.11 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr25_-_19648154 | 0.11 |
ENSDART00000148570
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr18_+_33171765 | 0.11 |
ENSDART00000064153
|
si:ch73-125k17.2
|
si:ch73-125k17.2 |
| chr13_-_41901225 | 0.10 |
ENSDART00000167582
|
ipmka
|
inositol polyphosphate multikinase a |
| chr4_-_16345227 | 0.10 |
ENSDART00000079521
|
kera
|
keratocan |
| chr15_-_17813680 | 0.10 |
ENSDART00000158556
|
CT573342.2
|
|
| chr7_+_20030888 | 0.10 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr7_+_48667081 | 0.10 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr11_+_2391469 | 0.10 |
ENSDART00000182121
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr20_-_7583486 | 0.10 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr12_-_990149 | 0.10 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr13_+_30228077 | 0.10 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr2_+_15203322 | 0.10 |
ENSDART00000144171
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr1_-_42289704 | 0.10 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr7_-_27685365 | 0.10 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr21_+_21817975 | 0.10 |
ENSDART00000151292
ENSDART00000166704 |
neu3.5
|
sialidase 3 (membrane sialidase), tandem duplicate 5 |
| chr14_-_899170 | 0.10 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr7_-_6467510 | 0.10 |
ENSDART00000166041
|
FP325123.1
|
Histone H3.2 |
| chr13_-_29505604 | 0.10 |
ENSDART00000110005
|
cdhr1a
|
cadherin-related family member 1a |
| chr21_+_42717424 | 0.10 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_+_54222156 | 0.10 |
ENSDART00000165201
ENSDART00000158518 |
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_-_12063381 | 0.10 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr6_+_39942417 | 0.10 |
ENSDART00000183256
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr8_-_43834442 | 0.10 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr21_+_43404945 | 0.10 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr20_-_27225876 | 0.10 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr4_-_16001118 | 0.10 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr22_-_10470663 | 0.10 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr12_+_16281312 | 0.10 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr11_-_45148743 | 0.10 |
ENSDART00000187124
ENSDART00000172878 ENSDART00000170993 |
cant1b
afmid
|
calcium activated nucleotidase 1b arylformamidase |
| chr5_-_30481263 | 0.10 |
ENSDART00000086734
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr5_-_23715861 | 0.10 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr6_+_43426599 | 0.10 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr20_+_48129759 | 0.09 |
ENSDART00000148494
|
tram2
|
translocation associated membrane protein 2 |
| chr7_-_48667056 | 0.09 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr23_+_20689255 | 0.09 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr16_-_16643377 | 0.09 |
ENSDART00000185748
|
tnxba
|
tenascin XBa |
| chr8_+_2575993 | 0.09 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr18_-_16801033 | 0.09 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr14_-_17068712 | 0.09 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr3_-_13921173 | 0.09 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr13_-_31017960 | 0.09 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr17_+_21486047 | 0.09 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr5_+_6672870 | 0.09 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr1_+_44003654 | 0.09 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr10_-_34089779 | 0.09 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr7_+_22586800 | 0.09 |
ENSDART00000035325
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr4_+_39742119 | 0.09 |
ENSDART00000176004
|
CR749167.2
|
|
| chr12_+_18663154 | 0.09 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr14_+_44794936 | 0.09 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr20_-_43775495 | 0.09 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr14_+_8715527 | 0.09 |
ENSDART00000170754
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr3_-_5555500 | 0.09 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr19_+_10339538 | 0.09 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr12_+_20412564 | 0.09 |
ENSDART00000186783
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr19_+_15440841 | 0.09 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr10_-_34200750 | 0.09 |
ENSDART00000137279
|
pimr148
|
Pim proto-oncogene, serine/threonine kinase, related 148 |
| chr2_+_50967947 | 0.08 |
ENSDART00000162288
|
si:ch211-249o11.5
|
si:ch211-249o11.5 |
| chr4_+_53268458 | 0.08 |
ENSDART00000165335
|
si:dkey-250k10.1
|
si:dkey-250k10.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prrx1a+prrx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 | 0.6 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 3.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.2 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.0 | 0.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2000648 | positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 3.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.6 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.4 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.2 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |