Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
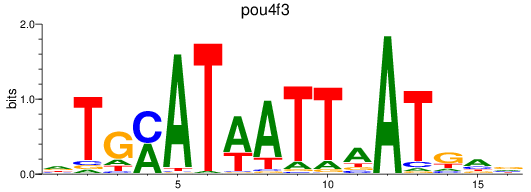
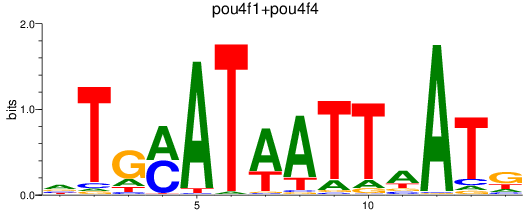
Results for pou4f3_pou4f1+pou4f4
Z-value: 3.28
Transcription factors associated with pou4f3_pou4f1+pou4f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f3
|
ENSDARG00000006206 | POU class 4 homeobox 3 |
|
pou4f1
|
ENSDARG00000005559 | POU class 4 homeobox 1 |
|
pou4f4
|
ENSDARG00000044375 | zgc |
|
pou4f4
|
ENSDARG00000111929 | zgc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f1 | dr11_v1_chr6_+_4539953_4539953 | 0.97 | 5.0e-03 | Click! |
| zgc:158291 | dr11_v1_chr21_+_38312549_38312549 | 0.90 | 3.7e-02 | Click! |
| pou4f3 | dr11_v1_chr9_+_54290896_54290896 | 0.75 | 1.4e-01 | Click! |
Activity profile of pou4f3_pou4f1+pou4f4 motif
Sorted Z-values of pou4f3_pou4f1+pou4f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_21688176 | 4.74 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr23_-_27345425 | 4.58 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr20_+_9355912 | 4.13 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr5_+_58372164 | 3.91 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr24_-_3419998 | 3.79 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr2_+_50608099 | 3.57 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr24_+_14713776 | 3.48 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr23_-_24195723 | 3.42 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr21_-_13123176 | 3.32 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_-_24195519 | 3.30 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr16_+_10318893 | 3.27 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr11_+_6819050 | 3.01 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr4_+_12111154 | 3.01 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr23_-_29376859 | 2.95 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr13_-_43599898 | 2.75 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr16_+_14029283 | 2.74 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr16_-_12953739 | 2.70 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr11_+_37137196 | 2.62 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr2_+_39108339 | 2.53 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr9_-_6661657 | 2.52 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr6_-_39489190 | 2.42 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr2_+_25929619 | 2.42 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr20_-_34801181 | 2.34 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr9_-_18735256 | 2.29 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr25_-_13614863 | 2.19 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr14_-_18672561 | 2.18 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr5_+_4054704 | 2.18 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr4_+_9669717 | 2.16 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr5_-_23429228 | 2.07 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr17_-_51893123 | 2.01 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr20_-_31808779 | 1.97 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr7_-_28148310 | 1.96 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr5_-_31689796 | 1.96 |
ENSDART00000184319
ENSDART00000190229 ENSDART00000186294 ENSDART00000170313 ENSDART00000147065 ENSDART00000134427 ENSDART00000098172 |
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr17_+_33999630 | 1.93 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr20_+_22045089 | 1.93 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr3_+_22578369 | 1.93 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr16_+_10776688 | 1.87 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr12_-_10705916 | 1.84 |
ENSDART00000164038
|
FO704786.1
|
|
| chr11_-_38083397 | 1.78 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr1_-_22861348 | 1.77 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr19_+_30662529 | 1.69 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr19_-_10196370 | 1.69 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr11_+_23957440 | 1.63 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr9_+_7030016 | 1.60 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr5_-_30418636 | 1.58 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr22_-_3564563 | 1.57 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr23_-_27692717 | 1.57 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr22_+_15507218 | 1.56 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr1_+_38758261 | 1.56 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr10_+_42423318 | 1.52 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr1_+_38758445 | 1.45 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr22_-_28226948 | 1.44 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr1_+_31113951 | 1.44 |
ENSDART00000129362
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr12_+_2446837 | 1.43 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr6_-_16394528 | 1.43 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr3_-_23407720 | 1.41 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr20_+_37661547 | 1.38 |
ENSDART00000007253
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr11_+_19060278 | 1.37 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr6_-_41229787 | 1.37 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr9_+_34319333 | 1.37 |
ENSDART00000035522
ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr6_-_35446110 | 1.37 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr1_-_15797663 | 1.37 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr20_-_44575103 | 1.36 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr11_+_23760470 | 1.35 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr7_+_73397283 | 1.35 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr20_+_41756996 | 1.33 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr11_+_25101220 | 1.33 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr19_+_37620342 | 1.31 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr7_-_69298271 | 1.30 |
ENSDART00000122935
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr20_+_27298783 | 1.30 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr5_+_70271799 | 1.25 |
ENSDART00000101316
|
znf618
|
zinc finger protein 618 |
| chr4_+_6869847 | 1.25 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr25_-_13614702 | 1.25 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr3_-_16055432 | 1.24 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr6_+_45494227 | 1.24 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr2_+_22702488 | 1.22 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr3_-_23406964 | 1.21 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr15_+_32297441 | 1.20 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr16_-_49505275 | 1.19 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr17_+_26208630 | 1.17 |
ENSDART00000087084
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr20_+_31076488 | 1.16 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr15_-_14552101 | 1.14 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_+_26352372 | 1.13 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr3_-_28665291 | 1.10 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr18_+_34362608 | 1.09 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr3_-_28048475 | 1.07 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr13_-_39830999 | 1.02 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr25_+_21833287 | 1.01 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr25_+_24291156 | 1.00 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr15_+_24704798 | 1.00 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr17_-_20287530 | 1.00 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr16_+_1353894 | 0.99 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr18_-_47662696 | 0.99 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr1_+_44491077 | 0.97 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr9_-_23147026 | 0.96 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr16_+_43401005 | 0.96 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr24_-_26622423 | 0.96 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr20_+_20638034 | 0.94 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr6_+_21740672 | 0.94 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr6_+_38427570 | 0.93 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr17_+_31914877 | 0.93 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr14_+_34495216 | 0.92 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr8_+_668184 | 0.91 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr20_+_17783404 | 0.91 |
ENSDART00000181946
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr13_+_27951688 | 0.91 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr2_-_9818640 | 0.90 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr21_-_39177564 | 0.90 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr19_+_42693855 | 0.89 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr3_-_61336841 | 0.89 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr8_+_7975745 | 0.89 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr10_+_40214877 | 0.86 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr11_+_30057762 | 0.86 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr13_+_23282549 | 0.85 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr24_+_3328354 | 0.85 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr9_+_44430705 | 0.84 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_+_25783801 | 0.84 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr10_+_38775959 | 0.83 |
ENSDART00000192990
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr9_+_16449398 | 0.83 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr23_+_6232895 | 0.82 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr8_-_39952727 | 0.82 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr7_+_12371061 | 0.81 |
ENSDART00000053860
|
saxo2
|
stabilizer of axonemal microtubules 2 |
| chr2_-_22286828 | 0.81 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr21_+_26389391 | 0.81 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr10_+_38775408 | 0.81 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr9_+_11034967 | 0.81 |
ENSDART00000152081
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr16_+_29630965 | 0.80 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr6_-_6423885 | 0.80 |
ENSDART00000092257
|
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr23_-_30431333 | 0.79 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr17_+_9009098 | 0.79 |
ENSDART00000180856
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr13_+_8604710 | 0.79 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr20_+_20750284 | 0.78 |
ENSDART00000181524
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr2_-_24348948 | 0.77 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr11_+_34523132 | 0.77 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr14_-_47849216 | 0.77 |
ENSDART00000192796
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_5422439 | 0.76 |
ENSDART00000055047
ENSDART00000142248 |
stk16
|
serine/threonine kinase 16 |
| chr14_+_36231126 | 0.76 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr5_-_41103583 | 0.75 |
ENSDART00000051070
ENSDART00000074781 |
golph3
|
golgi phosphoprotein 3 |
| chr13_+_12299997 | 0.73 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr11_+_19060079 | 0.72 |
ENSDART00000158123
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr25_-_4148719 | 0.72 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr17_+_15983247 | 0.72 |
ENSDART00000154275
|
clmn
|
calmin |
| chr13_+_36622100 | 0.72 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr10_-_27566481 | 0.72 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr22_-_32360464 | 0.71 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr8_+_30600386 | 0.69 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr7_+_7019911 | 0.68 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr5_-_26323137 | 0.67 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr22_-_17585618 | 0.67 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr3_+_33440615 | 0.67 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr2_+_22042745 | 0.66 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr18_-_23875370 | 0.65 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_30784198 | 0.64 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr11_+_19370447 | 0.63 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr9_+_11034314 | 0.62 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr2_-_24348642 | 0.62 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr15_-_11681653 | 0.62 |
ENSDART00000180160
|
fkrp
|
fukutin related protein |
| chr19_-_47455944 | 0.62 |
ENSDART00000190005
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr20_-_11178022 | 0.60 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr21_+_27278120 | 0.60 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr20_-_31075972 | 0.59 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr19_-_43552252 | 0.59 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr5_+_42393896 | 0.57 |
ENSDART00000189550
|
BX548073.13
|
|
| chr7_+_27059330 | 0.57 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr11_-_1956204 | 0.57 |
ENSDART00000185541
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr1_-_21901589 | 0.55 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr22_-_29906764 | 0.54 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr13_-_32635859 | 0.54 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr2_+_32846602 | 0.53 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr12_-_19151708 | 0.53 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr12_+_8168272 | 0.53 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr22_+_11153590 | 0.52 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr9_-_40931637 | 0.52 |
ENSDART00000165103
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr8_-_22838364 | 0.52 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr3_-_45250924 | 0.51 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr19_-_3574060 | 0.51 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr13_+_1944451 | 0.50 |
ENSDART00000125914
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr20_-_10120442 | 0.49 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr9_+_44431174 | 0.48 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_-_25895189 | 0.48 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr17_+_2162916 | 0.47 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr2_+_38742338 | 0.47 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr17_-_8673278 | 0.46 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr7_-_71829649 | 0.46 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr24_+_17142881 | 0.46 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr23_+_21479958 | 0.45 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr1_+_22851261 | 0.45 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr13_-_30700460 | 0.45 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr20_-_2361226 | 0.44 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr18_+_40355408 | 0.44 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_+_30610547 | 0.43 |
ENSDART00000122256
|
ccr6a
|
chemokine (C-C motif) receptor 6a |
| chr1_-_44581937 | 0.43 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr17_+_15983557 | 0.43 |
ENSDART00000190806
|
clmn
|
calmin |
| chr20_-_39103119 | 0.42 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr17_-_8673567 | 0.42 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr15_-_14469704 | 0.41 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_45960191 | 0.41 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr4_-_9173552 | 0.41 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr20_-_40766387 | 0.40 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr7_+_6941583 | 0.39 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr20_-_6196989 | 0.39 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f3_pou4f1+pou4f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.0 | 3.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.7 | 2.9 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.7 | 6.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.5 | 1.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 0.9 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 0.9 | GO:0090247 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.3 | 0.8 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.3 | 1.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 4.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 0.7 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.2 | 3.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 2.6 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 1.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 2.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 2.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 1.9 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.7 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.9 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.5 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.9 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.6 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.8 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 3.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 1.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.1 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 1.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.0 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.3 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.0 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 2.0 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 1.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 2.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 5.2 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.5 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 2.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 1.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 2.4 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.5 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 2.2 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 3.2 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 7.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 3.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 0.8 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 6.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.3 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 2.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.5 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.6 | 1.9 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.4 | 8.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 2.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 2.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 2.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 8.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.1 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 3.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 3.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 5.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.4 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 1.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |