Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
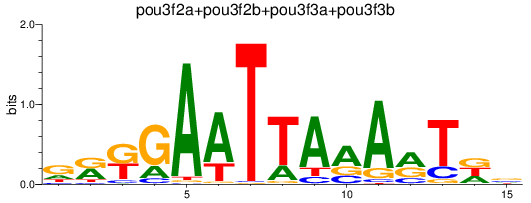
Results for pou3f2a+pou3f2b+pou3f3a+pou3f3b
Z-value: 1.56
Transcription factors associated with pou3f2a+pou3f2b+pou3f3a+pou3f3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f3a
|
ENSDARG00000042032 | POU class 3 homeobox 3a |
|
pou3f2a
|
ENSDARG00000070220 | POU class 3 homeobox 2a |
|
pou3f2b
|
ENSDARG00000076262 | POU class 3 homeobox 2b |
|
pou3f3b
|
ENSDARG00000095896 | POU class 3 homeobox 3b |
|
pou3f2b
|
ENSDARG00000112713 | POU class 3 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f2a | dr11_v1_chr4_+_5741733_5741733 | -0.35 | 5.6e-01 | Click! |
| pou3f3a | dr11_v1_chr9_-_6661657_6661666 | -0.33 | 5.9e-01 | Click! |
| pou3f3b | dr11_v1_chr6_+_14949950_14949950 | -0.33 | 5.9e-01 | Click! |
| pou3f2b | dr11_v1_chr16_+_32559821_32559821 | -0.26 | 6.7e-01 | Click! |
Activity profile of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
Sorted Z-values of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_25418856 | 0.83 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr7_-_16562200 | 0.75 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr19_-_5380770 | 0.64 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr20_-_29482492 | 0.63 |
ENSDART00000178308
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr23_-_24263474 | 0.55 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr3_-_43770876 | 0.51 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr4_-_67799941 | 0.46 |
ENSDART00000185830
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr1_+_53945934 | 0.45 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr25_-_27819838 | 0.44 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr10_-_42237304 | 0.43 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr16_+_23487051 | 0.42 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr18_+_8917766 | 0.42 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr2_-_23768818 | 0.41 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr24_-_36301072 | 0.40 |
ENSDART00000062736
|
coasy
|
CoA synthase |
| chr17_-_19345521 | 0.40 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr16_-_41990421 | 0.38 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr8_-_32497581 | 0.37 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr2_-_3419890 | 0.36 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr16_-_42523744 | 0.35 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr23_+_5631381 | 0.35 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr22_-_31020690 | 0.35 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr8_+_21353878 | 0.34 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr23_-_31372639 | 0.33 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr25_+_5035343 | 0.32 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr7_-_41851605 | 0.32 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr15_-_29598679 | 0.31 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr24_+_25471196 | 0.31 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr24_+_20920563 | 0.30 |
ENSDART00000037224
|
cst14a.2
|
cystatin 14a, tandem duplicate 2 |
| chr24_-_3426620 | 0.29 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr16_-_26140768 | 0.29 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr14_+_36889893 | 0.29 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr14_+_1007169 | 0.29 |
ENSDART00000172676
|
f8
|
coagulation factor VIII, procoagulant component |
| chr16_+_29516098 | 0.28 |
ENSDART00000174895
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr23_+_36653376 | 0.28 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr21_-_2814709 | 0.28 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr14_+_15620640 | 0.28 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr21_-_44565862 | 0.28 |
ENSDART00000180158
|
fundc2
|
fun14 domain containing 2 |
| chr15_+_963292 | 0.28 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr7_+_8367929 | 0.28 |
ENSDART00000173303
|
zgc:172143
|
zgc:172143 |
| chr7_-_8416750 | 0.27 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr18_-_26894732 | 0.27 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr7_-_8408014 | 0.27 |
ENSDART00000112492
|
zgc:194686
|
zgc:194686 |
| chr16_+_23796612 | 0.27 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr21_+_11401247 | 0.27 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr14_+_30340251 | 0.27 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr7_+_48761875 | 0.26 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr7_+_8370076 | 0.26 |
ENSDART00000057103
|
zgc:172143
|
zgc:172143 |
| chr10_+_15088534 | 0.26 |
ENSDART00000142865
|
si:ch211-95j8.3
|
si:ch211-95j8.3 |
| chr12_-_33746111 | 0.26 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr24_-_12958668 | 0.26 |
ENSDART00000178982
|
FITM1 (1 of many)
|
Danio rerio fat storage inducing transmembrane protein 1 (LOC792443), mRNA. |
| chr9_-_34509997 | 0.25 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr25_+_1549838 | 0.25 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr16_+_10776688 | 0.25 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr2_+_15048410 | 0.25 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr25_+_15647750 | 0.25 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr5_+_37091626 | 0.25 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr25_+_19238175 | 0.25 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr5_-_33259079 | 0.25 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr18_+_7591381 | 0.25 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr13_+_51956397 | 0.25 |
ENSDART00000161098
|
lclat1
|
lysocardiolipin acyltransferase 1 |
| chr3_-_32596394 | 0.24 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr7_+_72003301 | 0.24 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr7_-_62244744 | 0.24 |
ENSDART00000192522
ENSDART00000170635 ENSDART00000073872 |
CCKAR
|
cholecystokinin A receptor |
| chr5_-_30715225 | 0.24 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr7_-_19923249 | 0.24 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr20_+_30939178 | 0.24 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr16_+_10777116 | 0.23 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr8_-_36469117 | 0.23 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr11_-_41996957 | 0.23 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr25_-_13201458 | 0.23 |
ENSDART00000192451
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr16_-_26525901 | 0.23 |
ENSDART00000110260
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr1_-_23308225 | 0.22 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr7_+_22823889 | 0.22 |
ENSDART00000127467
ENSDART00000148576 ENSDART00000149993 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr13_-_33362191 | 0.22 |
ENSDART00000100514
|
zgc:172120
|
zgc:172120 |
| chr7_+_20109968 | 0.22 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr18_-_40773413 | 0.21 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr1_-_44710937 | 0.21 |
ENSDART00000139036
|
si:dkey-28b4.7
|
si:dkey-28b4.7 |
| chr4_-_8043839 | 0.21 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr6_-_32411703 | 0.21 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr11_+_39928828 | 0.21 |
ENSDART00000137516
ENSDART00000134082 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr19_+_26923110 | 0.21 |
ENSDART00000149988
|
nelfe
|
negative elongation factor complex member E |
| chr8_-_21142550 | 0.21 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr3_+_1942219 | 0.21 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr16_+_35916371 | 0.21 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr16_+_7653905 | 0.21 |
ENSDART00000081431
|
popdc3
|
popeye domain containing 3 |
| chr21_-_42831033 | 0.21 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr18_-_48296793 | 0.20 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr21_-_22317920 | 0.20 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr4_-_16412084 | 0.20 |
ENSDART00000188460
|
dcn
|
decorin |
| chr17_+_6538733 | 0.20 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr11_+_6080528 | 0.20 |
ENSDART00000183099
|
ushbp1
|
Usher syndrome 1C binding protein 1 |
| chr11_+_4026229 | 0.20 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr19_-_15229421 | 0.20 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr22_+_10651726 | 0.20 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr19_+_2631565 | 0.20 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr7_+_25221757 | 0.19 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr25_+_11496079 | 0.19 |
ENSDART00000163746
|
AGBL1
|
si:ch73-141f14.1 |
| chr5_-_65037525 | 0.19 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr6_-_39037613 | 0.19 |
ENSDART00000098906
|
tns2b
|
tensin 2b |
| chr1_-_9885320 | 0.19 |
ENSDART00000054843
ENSDART00000138630 |
zgc:56409
|
zgc:56409 |
| chr15_-_8665662 | 0.19 |
ENSDART00000090675
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr8_+_22485956 | 0.19 |
ENSDART00000099960
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr3_-_26921475 | 0.19 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr3_+_55093581 | 0.19 |
ENSDART00000101709
|
hbaa2
|
hemoglobin, alpha adult 2 |
| chr7_+_20110336 | 0.19 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr6_-_25163722 | 0.19 |
ENSDART00000192225
|
znf326
|
zinc finger protein 326 |
| chr18_-_46258612 | 0.19 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr21_-_22892124 | 0.19 |
ENSDART00000065563
|
ccdc90b
|
coiled-coil domain containing 90B |
| chr6_+_58607488 | 0.19 |
ENSDART00000182648
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr15_-_17813680 | 0.19 |
ENSDART00000158556
|
CT573342.2
|
|
| chr6_+_2093206 | 0.19 |
ENSDART00000114314
|
tgm2b
|
transglutaminase 2b |
| chr13_-_15986871 | 0.19 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr6_-_15101477 | 0.19 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr1_-_7998493 | 0.19 |
ENSDART00000185901
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr16_-_38001040 | 0.18 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr15_+_29116063 | 0.18 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr4_-_9214288 | 0.18 |
ENSDART00000067309
|
avpr1ab
|
arginine vasopressin receptor 1Ab |
| chr10_-_35220285 | 0.18 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr23_-_31436524 | 0.18 |
ENSDART00000140519
|
zgc:153284
|
zgc:153284 |
| chr11_+_36243774 | 0.18 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr4_+_18843015 | 0.18 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr16_+_41826584 | 0.18 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr6_-_43047774 | 0.18 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr6_+_2093367 | 0.18 |
ENSDART00000148396
|
tgm2b
|
transglutaminase 2b |
| chr4_+_77657660 | 0.18 |
ENSDART00000099744
|
si:dkey-61p9.7
|
si:dkey-61p9.7 |
| chr2_-_5135125 | 0.18 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr5_-_65037371 | 0.18 |
ENSDART00000170560
|
anxa1b
|
annexin A1b |
| chr18_-_12295092 | 0.18 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr13_+_31757331 | 0.18 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr11_-_863832 | 0.18 |
ENSDART00000173229
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr15_-_43284021 | 0.18 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr24_+_3963684 | 0.17 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr2_-_55337585 | 0.17 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr21_-_31252131 | 0.17 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr6_-_15639087 | 0.17 |
ENSDART00000128940
ENSDART00000183092 |
mlpha
|
melanophilin a |
| chr19_-_3781405 | 0.17 |
ENSDART00000170609
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr5_+_24087035 | 0.17 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr5_-_55560937 | 0.17 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr5_-_4418555 | 0.17 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr1_-_8000428 | 0.17 |
ENSDART00000133098
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr23_+_17839187 | 0.17 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr24_+_11381400 | 0.17 |
ENSDART00000058703
|
ackr4b
|
atypical chemokine receptor 4b |
| chr18_-_12327426 | 0.17 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr3_+_49021079 | 0.17 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr24_+_6353394 | 0.17 |
ENSDART00000165118
|
CR352329.1
|
|
| chr8_-_49766205 | 0.17 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr21_-_38730557 | 0.17 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr3_+_5083407 | 0.17 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr13_-_22766445 | 0.17 |
ENSDART00000140151
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr19_+_42071033 | 0.17 |
ENSDART00000169067
ENSDART00000183436 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr5_+_30384554 | 0.17 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr22_-_24297510 | 0.17 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr10_+_15187316 | 0.16 |
ENSDART00000179732
|
CR925756.1
|
|
| chr12_+_15690363 | 0.16 |
ENSDART00000166803
ENSDART00000184178 ENSDART00000161500 ENSDART00000170017 |
si:dkeyp-94b4.1
|
si:dkeyp-94b4.1 |
| chr6_+_584632 | 0.16 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr12_+_34051848 | 0.16 |
ENSDART00000153276
|
cyth1b
|
cytohesin 1b |
| chr21_-_15674802 | 0.16 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr10_-_2788668 | 0.16 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr12_-_37653685 | 0.16 |
ENSDART00000085121
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr20_+_28364742 | 0.16 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr14_+_6953244 | 0.16 |
ENSDART00000159470
|
rack1
|
receptor for activated C kinase 1 |
| chr22_+_19553390 | 0.16 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr3_+_17933132 | 0.16 |
ENSDART00000104299
ENSDART00000162144 ENSDART00000162242 ENSDART00000166289 ENSDART00000171101 ENSDART00000164853 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr18_-_7032227 | 0.16 |
ENSDART00000127138
|
calub
|
calumenin b |
| chr25_-_8660726 | 0.16 |
ENSDART00000168010
ENSDART00000171709 ENSDART00000163426 |
unc45a
|
unc-45 myosin chaperone A |
| chr16_+_20167811 | 0.16 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr2_+_36121373 | 0.16 |
ENSDART00000187002
|
CT867973.2
|
|
| chr4_-_13931508 | 0.15 |
ENSDART00000067174
|
zcrb1
|
zinc finger CCHC-type and RNA binding motif 1 |
| chr17_+_5061135 | 0.15 |
ENSDART00000064313
|
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr23_+_9268083 | 0.15 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr5_+_27488975 | 0.15 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr3_-_34069637 | 0.15 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr2_-_44199722 | 0.15 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr11_-_40519886 | 0.15 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr2_+_35595454 | 0.15 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr16_-_10316359 | 0.15 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr15_+_34699585 | 0.15 |
ENSDART00000017569
|
tnfb
|
tumor necrosis factor b (TNF superfamily, member 2) |
| chr25_-_37084032 | 0.15 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr17_-_28707898 | 0.15 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr9_+_28688574 | 0.15 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr20_+_45620076 | 0.15 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr22_-_14161309 | 0.15 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr1_+_29862074 | 0.15 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr23_+_35847200 | 0.14 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr25_-_3836597 | 0.14 |
ENSDART00000115154
|
si:ch211-247i17.1
|
si:ch211-247i17.1 |
| chr9_-_695000 | 0.14 |
ENSDART00000181318
ENSDART00000115030 |
dip2a
|
disco-interacting protein 2 homolog A |
| chr2_-_14656669 | 0.14 |
ENSDART00000190219
|
CABZ01026681.1
|
|
| chr13_+_35925490 | 0.14 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr10_-_3138403 | 0.14 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr24_-_8777781 | 0.14 |
ENSDART00000082362
ENSDART00000177400 |
mak
|
male germ cell-associated kinase |
| chr7_+_8361083 | 0.14 |
ENSDART00000102091
|
jac7
|
jacalin 7 |
| chr1_+_44767657 | 0.14 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chr1_-_22687913 | 0.14 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr16_-_2650341 | 0.14 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr9_+_30421489 | 0.14 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr11_+_43661735 | 0.14 |
ENSDART00000017912
|
gli2b
|
GLI family zinc finger 2b |
| chr5_-_33959868 | 0.14 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr5_+_29794058 | 0.14 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr15_+_1134870 | 0.14 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr7_+_31132588 | 0.14 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f2a+pou3f2b+pou3f3a+pou3f3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 0.7 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 0.4 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.5 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.2 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.9 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.2 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.1 | 0.2 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.0 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.3 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.4 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0048855 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0035142 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.0 | 0.2 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.1 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.0 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0045622 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T cell differentiation(GO:0045581) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.0 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.0 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.2 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |