Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
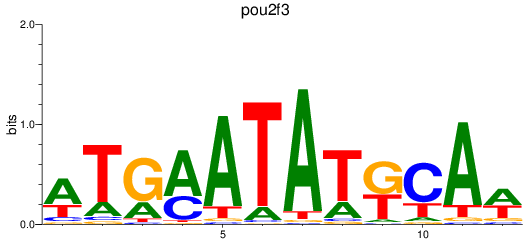
Results for pou2f3
Z-value: 1.39
Transcription factors associated with pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f3
|
ENSDARG00000052387 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f3 | dr11_v1_chr5_+_58550291_58550291 | 0.08 | 9.0e-01 | Click! |
Activity profile of pou2f3 motif
Sorted Z-values of pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_22476742 | 1.33 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr18_-_20869175 | 1.28 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr6_+_6924637 | 1.16 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr25_+_3058924 | 0.96 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr1_+_25801648 | 0.79 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr10_+_39084354 | 0.78 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr23_+_22200467 | 0.71 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr4_+_4509996 | 0.67 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr20_+_572037 | 0.59 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr19_-_7450796 | 0.51 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr6_-_20875111 | 0.51 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr3_+_39566999 | 0.51 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr11_+_37275448 | 0.51 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr13_-_2189761 | 0.48 |
ENSDART00000166255
|
mlip
|
muscular LMNA-interacting protein |
| chr25_+_35019693 | 0.47 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr24_+_792429 | 0.46 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr13_-_2215213 | 0.46 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr23_-_35066816 | 0.45 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr5_-_25583125 | 0.44 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr20_-_43750771 | 0.44 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr1_+_51039558 | 0.43 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr16_-_42523744 | 0.43 |
ENSDART00000017185
|
tbx20
|
T-box 20 |
| chr5_-_25582721 | 0.42 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr19_-_25114701 | 0.42 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_+_35189555 | 0.42 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr1_-_38815361 | 0.41 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr11_-_36341028 | 0.41 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr25_+_14017609 | 0.40 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr10_-_22918214 | 0.39 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr7_-_18168493 | 0.39 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr11_-_36341189 | 0.39 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr3_-_50865079 | 0.38 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr25_-_13490744 | 0.35 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr15_-_43284021 | 0.35 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr2_-_48171112 | 0.35 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr22_-_26595027 | 0.35 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr7_-_58098814 | 0.34 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr4_+_7391110 | 0.34 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr13_-_30645965 | 0.34 |
ENSDART00000109307
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr10_+_5268054 | 0.34 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr21_-_37733287 | 0.33 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr18_-_12327426 | 0.33 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr8_+_49433663 | 0.32 |
ENSDART00000140481
ENSDART00000180714 |
nfu1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr20_+_23625387 | 0.32 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr16_-_5721386 | 0.32 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr24_-_7321928 | 0.32 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr17_+_24109012 | 0.32 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr2_-_3419890 | 0.31 |
ENSDART00000055618
|
iba57
|
IBA57, iron-sulfur cluster assembly |
| chr21_+_723998 | 0.31 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr17_-_26926577 | 0.30 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr10_+_2899108 | 0.30 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr9_-_1702648 | 0.30 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr10_+_35439952 | 0.30 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr3_-_59981476 | 0.30 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr18_+_16125852 | 0.29 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr24_-_28333029 | 0.29 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr8_-_14052349 | 0.29 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr22_+_14051894 | 0.28 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr3_-_59981162 | 0.28 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_+_40150612 | 0.28 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr22_-_19552796 | 0.27 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr14_-_45967981 | 0.27 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr16_+_2820340 | 0.27 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr22_-_11438627 | 0.27 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr6_-_30932078 | 0.27 |
ENSDART00000028612
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr20_+_29691118 | 0.26 |
ENSDART00000164121
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr17_-_29213710 | 0.26 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr4_+_12292274 | 0.26 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr9_-_21230999 | 0.26 |
ENSDART00000150160
ENSDART00000102147 |
pla1a
|
phospholipase A1 member A |
| chr19_-_205104 | 0.26 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr18_+_2222447 | 0.26 |
ENSDART00000185927
|
pigbos1
|
PIGB opposite strand 1 |
| chr23_+_33907899 | 0.26 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr4_+_25950372 | 0.26 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr11_-_44485700 | 0.25 |
ENSDART00000186613
ENSDART00000172828 ENSDART00000113222 |
mfn1b
|
mitofusin 1b |
| chr17_+_21964472 | 0.25 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr4_-_12388535 | 0.25 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr5_+_39504136 | 0.25 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr1_-_36151377 | 0.25 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr5_+_22970617 | 0.24 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr2_-_42558549 | 0.24 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr25_+_16601839 | 0.24 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr18_-_2222128 | 0.24 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr25_+_31277415 | 0.24 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr13_+_33282095 | 0.24 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr4_-_390431 | 0.24 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr4_-_15603511 | 0.23 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr1_-_12278056 | 0.23 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr11_+_30647545 | 0.23 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr19_-_22387141 | 0.23 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr3_-_39305291 | 0.23 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr17_-_30975707 | 0.22 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr7_+_55633483 | 0.22 |
ENSDART00000180993
ENSDART00000184845 |
trappc2l
|
trafficking protein particle complex 2-like |
| chr9_-_7212973 | 0.22 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr21_-_40717678 | 0.22 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr7_-_44704910 | 0.22 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr11_-_43200994 | 0.21 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_-_24659345 | 0.21 |
ENSDART00000128902
ENSDART00000190684 |
proser1
|
proline and serine rich 1 |
| chr19_-_31686252 | 0.21 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr22_+_18389271 | 0.21 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr3_-_5067585 | 0.21 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr1_-_51720633 | 0.20 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr18_+_30567945 | 0.20 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr4_-_9579299 | 0.20 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr3_+_59851537 | 0.20 |
ENSDART00000180997
|
CU693479.1
|
|
| chr10_-_42751641 | 0.20 |
ENSDART00000182734
ENSDART00000113926 |
zgc:100918
|
zgc:100918 |
| chr11_-_762721 | 0.20 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr1_+_51496862 | 0.20 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr21_+_34119759 | 0.20 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr12_+_17504559 | 0.20 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr18_+_38885309 | 0.20 |
ENSDART00000041597
|
arpp19a
|
cAMP-regulated phosphoprotein 19a |
| chr5_-_65662996 | 0.20 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr24_-_21989406 | 0.20 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr19_+_5076617 | 0.20 |
ENSDART00000189990
|
eno2
|
enolase 2 |
| chr16_+_46111849 | 0.20 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr20_+_29690901 | 0.19 |
ENSDART00000142669
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr10_+_23060391 | 0.19 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr24_+_39614853 | 0.19 |
ENSDART00000165138
|
BX547934.2
|
|
| chr11_+_13176568 | 0.19 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr8_+_22931427 | 0.19 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr5_-_32274383 | 0.19 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr1_-_36152131 | 0.19 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr9_+_54179306 | 0.19 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr1_-_53756851 | 0.19 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr16_+_4531730 | 0.19 |
ENSDART00000122861
|
tfpt
|
TCF3 (E2A) fusion partner |
| chr21_+_34088377 | 0.18 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr19_-_5103313 | 0.18 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_-_18410968 | 0.18 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr2_+_6243144 | 0.18 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr19_+_33553586 | 0.18 |
ENSDART00000183477
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr25_+_19238175 | 0.18 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr20_-_31497300 | 0.17 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr25_+_6186823 | 0.17 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr13_-_44423 | 0.17 |
ENSDART00000093247
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr1_-_26063188 | 0.17 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr14_-_17588345 | 0.17 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr9_-_30165621 | 0.17 |
ENSDART00000089543
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr1_-_41287914 | 0.17 |
ENSDART00000022546
ENSDART00000126853 ENSDART00000125988 |
rgs12b
|
regulator of G protein signaling 12b |
| chr17_-_25649079 | 0.17 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr2_-_37632896 | 0.17 |
ENSDART00000008302
|
insra
|
insulin receptor a |
| chr17_-_37395460 | 0.17 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr2_-_9607879 | 0.17 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr5_-_54481692 | 0.17 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr5_+_24245682 | 0.17 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr17_-_6451801 | 0.17 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr9_+_12444494 | 0.16 |
ENSDART00000102430
|
tmem41aa
|
transmembrane protein 41aa |
| chr8_-_43158486 | 0.16 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr15_-_43327911 | 0.16 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr5_+_64732036 | 0.16 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr21_-_3613702 | 0.16 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr25_-_23526058 | 0.16 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr18_-_15551360 | 0.16 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr25_-_20666328 | 0.15 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr4_-_1801519 | 0.15 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr7_+_30725473 | 0.15 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr17_-_30975978 | 0.15 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr23_-_26111760 | 0.15 |
ENSDART00000164032
|
rca2.1
|
regulator of complement activation group 2 gene 1 |
| chr20_-_3087162 | 0.15 |
ENSDART00000152495
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr14_-_33297287 | 0.15 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr16_-_42390441 | 0.15 |
ENSDART00000148475
|
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr13_+_45431660 | 0.15 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr22_+_28446557 | 0.15 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr24_+_39186940 | 0.15 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr16_-_44349845 | 0.15 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr21_-_38730557 | 0.15 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr14_-_4044545 | 0.15 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr20_+_30578967 | 0.15 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr2_+_5563077 | 0.14 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr4_-_2052687 | 0.14 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr7_+_19738665 | 0.14 |
ENSDART00000089333
|
tkfc
|
triokinase/FMN cyclase |
| chr7_-_66877058 | 0.14 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr23_-_33709964 | 0.14 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr15_+_1134870 | 0.14 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr9_+_34319333 | 0.14 |
ENSDART00000035522
ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr5_+_51848756 | 0.14 |
ENSDART00000087467
ENSDART00000184466 |
cmya5
|
cardiomyopathy associated 5 |
| chr25_+_31227747 | 0.14 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr10_+_18911152 | 0.14 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr19_+_1510971 | 0.14 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr12_+_3571770 | 0.13 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr24_-_24849091 | 0.13 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr19_-_25772980 | 0.13 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr20_-_54198130 | 0.13 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr25_-_3058687 | 0.13 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr12_+_18543783 | 0.13 |
ENSDART00000148326
ENSDART00000134530 |
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr2_+_51645164 | 0.13 |
ENSDART00000169600
|
abhd4
|
abhydrolase domain containing 4 |
| chr9_-_54840124 | 0.13 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr8_-_28274251 | 0.13 |
ENSDART00000050671
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr9_-_32343673 | 0.13 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr14_-_33894915 | 0.13 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr2_-_1364678 | 0.13 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr25_+_5012791 | 0.13 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr12_-_19862912 | 0.13 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr25_-_25058508 | 0.13 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr21_-_34032650 | 0.13 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr10_-_27566481 | 0.12 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr17_-_49438873 | 0.12 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr24_+_5208171 | 0.12 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr17_-_50490475 | 0.12 |
ENSDART00000075159
|
zgc:113886
|
zgc:113886 |
| chr14_-_30876299 | 0.12 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr3_-_58798815 | 0.12 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr16_+_4695075 | 0.12 |
ENSDART00000039054
|
mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.9 | GO:0032623 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 1.0 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.5 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.2 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.4 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 0.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.2 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.2 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 0.1 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.0 | 0.3 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.0 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.8 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.0 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.5 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.7 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.6 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |