Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
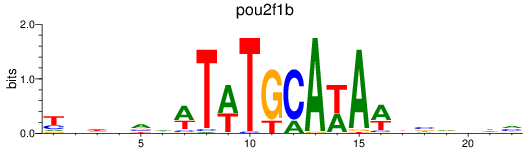
Results for pou2f1b
Z-value: 1.79
Transcription factors associated with pou2f1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f1b
|
ENSDARG00000007996 | POU class 2 homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f1b | dr11_v1_chr9_+_34319333_34319333 | -0.81 | 9.3e-02 | Click! |
Activity profile of pou2f1b motif
Sorted Z-values of pou2f1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_35458190 | 1.61 |
ENSDART00000051313
|
fbp1b
|
fructose-1,6-bisphosphatase 1b |
| chr1_+_52735484 | 1.60 |
ENSDART00000182076
|
CABZ01021532.1
|
|
| chr24_+_23959487 | 1.28 |
ENSDART00000080487
|
hgd
|
homogentisate 1,2-dioxygenase |
| chr19_+_2835240 | 1.22 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr4_-_8043839 | 1.20 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr25_-_18454016 | 1.20 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr20_+_10538025 | 1.19 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr10_+_2669405 | 1.15 |
ENSDART00000110202
|
ccl27b
|
chemokine (C-C motif) ligand 27b |
| chr24_-_23758003 | 1.14 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr1_-_56080112 | 1.04 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr3_-_45326897 | 1.01 |
ENSDART00000086417
|
il21r.2
|
interleukin 21 receptor, tandem duplicate 2 |
| chr9_-_33063083 | 0.99 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr10_+_26667475 | 0.99 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr20_+_10544100 | 0.97 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_4145594 | 0.97 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr24_-_21150100 | 0.96 |
ENSDART00000154249
ENSDART00000193256 |
gramd1c
|
GRAM domain containing 1c |
| chr14_-_46198373 | 0.88 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr3_+_37112693 | 0.85 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr4_-_8035520 | 0.85 |
ENSDART00000146146
|
si:ch211-240l19.7
|
si:ch211-240l19.7 |
| chr20_-_35750810 | 0.84 |
ENSDART00000153072
|
adgrf8
|
adhesion G protein-coupled receptor F8 |
| chr2_+_27394798 | 0.84 |
ENSDART00000115071
|
selenop2
|
selenoprotein P2 |
| chr9_-_33062891 | 0.84 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr2_-_20120904 | 0.83 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr18_-_6742287 | 0.82 |
ENSDART00000140752
|
miox
|
myo-inositol oxygenase |
| chr5_+_39087364 | 0.80 |
ENSDART00000004286
|
anxa3a
|
annexin A3a |
| chr2_+_49457449 | 0.79 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr2_-_51757328 | 0.78 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr11_-_25418856 | 0.78 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr17_+_2549503 | 0.75 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr25_-_13029680 | 0.74 |
ENSDART00000166151
|
ccl39.2
|
chemokine (C-C motif) ligand 39, duplicate 2 |
| chr16_+_38394371 | 0.73 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr17_+_23311377 | 0.72 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr2_+_31665836 | 0.71 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr11_+_23993298 | 0.70 |
ENSDART00000186757
ENSDART00000172459 |
chia.1
|
chitinase, acidic.1 |
| chr7_-_12968689 | 0.70 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr20_+_52492192 | 0.70 |
ENSDART00000057986
|
TSTA3 (1 of many)
|
zgc:100864 |
| chr11_-_2478374 | 0.69 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr12_-_4243268 | 0.68 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr16_-_23346095 | 0.67 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr19_-_977849 | 0.67 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr17_+_28102487 | 0.67 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr23_-_33775145 | 0.67 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr1_-_10071422 | 0.66 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr18_+_39060687 | 0.65 |
ENSDART00000098729
ENSDART00000136367 |
zgc:171509
|
zgc:171509 |
| chr1_+_58922027 | 0.64 |
ENSDART00000159479
|
trip10b
|
thyroid hormone receptor interactor 10b |
| chr5_+_35458346 | 0.64 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr6_-_442163 | 0.63 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr9_-_34396264 | 0.63 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr2_-_21438492 | 0.63 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr14_-_21064199 | 0.62 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr10_-_244745 | 0.61 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr18_+_44703343 | 0.61 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr5_+_11840905 | 0.60 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr14_+_10461264 | 0.60 |
ENSDART00000081095
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr4_+_1461079 | 0.60 |
ENSDART00000082383
|
trim35-14
|
tripartite motif containing 35-14 |
| chr10_-_17466990 | 0.60 |
ENSDART00000147794
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr3_+_12816362 | 0.59 |
ENSDART00000163743
ENSDART00000170788 |
cyp2k6
|
cytochrome P450, family 2, subfamily K, polypeptide 6 |
| chr1_-_7582859 | 0.59 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr2_+_37875789 | 0.59 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr2_+_42191592 | 0.59 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_+_49043917 | 0.59 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr25_+_186583 | 0.58 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr24_-_36680261 | 0.58 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr21_-_34926619 | 0.57 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr1_+_9153141 | 0.57 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr12_-_17655683 | 0.57 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr22_-_23706771 | 0.56 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr15_+_32711663 | 0.56 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr20_-_49889111 | 0.56 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr23_+_39695827 | 0.55 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr3_-_4552590 | 0.54 |
ENSDART00000043148
ENSDART00000132224 |
ftr43
|
finTRIM family, member 43 |
| chr1_+_7546259 | 0.53 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr20_-_29499363 | 0.53 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_49021079 | 0.53 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr19_-_27570333 | 0.53 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr18_-_29925717 | 0.53 |
ENSDART00000099281
|
mhc2dbb
|
major histocompatibility complex class II DBB gene |
| chr17_+_22530820 | 0.52 |
ENSDART00000089916
|
edaradd
|
EDAR-associated death domain |
| chr1_-_23274393 | 0.52 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr24_+_7322116 | 0.52 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr4_+_7822773 | 0.52 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr10_+_38643304 | 0.51 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr11_-_29658396 | 0.51 |
ENSDART00000183947
|
rpl22
|
ribosomal protein L22 |
| chr15_-_23814330 | 0.51 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr5_+_26799165 | 0.49 |
ENSDART00000145736
|
tcn2
|
transcobalamin II |
| chr15_-_18209672 | 0.49 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr15_-_37767901 | 0.49 |
ENSDART00000154763
|
si:dkey-42l23.4
|
si:dkey-42l23.4 |
| chr19_-_340641 | 0.49 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr8_+_23436411 | 0.48 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr18_+_15758375 | 0.47 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr20_-_53624645 | 0.47 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr16_+_19637384 | 0.47 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr10_+_34001444 | 0.46 |
ENSDART00000149934
|
kl
|
klotho |
| chr21_+_15295685 | 0.46 |
ENSDART00000135603
|
BX901878.1
|
|
| chr11_-_21404044 | 0.45 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr19_+_23301050 | 0.45 |
ENSDART00000193713
|
si:dkey-79i2.4
|
si:dkey-79i2.4 |
| chr17_+_16046314 | 0.45 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_+_5341592 | 0.44 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr22_+_29994093 | 0.44 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr7_-_51102479 | 0.44 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr15_+_14856307 | 0.44 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr1_-_55262763 | 0.44 |
ENSDART00000152769
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr3_-_61205711 | 0.43 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr1_-_44638058 | 0.43 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr10_-_35257458 | 0.43 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr5_-_11809710 | 0.43 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr13_+_30228077 | 0.43 |
ENSDART00000100813
ENSDART00000147729 ENSDART00000133404 |
rps24
|
ribosomal protein S24 |
| chr22_-_23668356 | 0.43 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr17_-_38291065 | 0.43 |
ENSDART00000152056
|
pax9
|
paired box 9 |
| chr20_-_35578435 | 0.43 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr20_-_41992878 | 0.42 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr9_-_12443726 | 0.42 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr17_+_21486047 | 0.42 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr4_+_5249494 | 0.42 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr18_-_26715655 | 0.41 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr23_+_7692042 | 0.41 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr20_-_45812144 | 0.41 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr19_-_27550768 | 0.41 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr20_+_4221978 | 0.41 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_5070794 | 0.41 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr18_-_26715156 | 0.41 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr10_-_40819022 | 0.41 |
ENSDART00000076316
ENSDART00000141329 |
zgc:113625
|
zgc:113625 |
| chr21_+_25625026 | 0.40 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr20_+_18993452 | 0.40 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr20_-_51307815 | 0.40 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr17_-_6618574 | 0.40 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr14_-_7409364 | 0.40 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr24_+_12074340 | 0.40 |
ENSDART00000170997
ENSDART00000166227 |
ccr9b
|
chemokine (C-C motif) receptor 9b |
| chr16_+_50289916 | 0.39 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr5_+_29714786 | 0.39 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr4_-_77602685 | 0.39 |
ENSDART00000152922
|
si:dkey-61p9.11
|
si:dkey-61p9.11 |
| chr25_+_22320738 | 0.38 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr14_+_36886950 | 0.38 |
ENSDART00000183719
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr4_+_76705830 | 0.38 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr5_+_29715040 | 0.38 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr25_+_13205878 | 0.38 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr19_+_30990815 | 0.38 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr4_-_49312747 | 0.38 |
ENSDART00000181057
|
BX942819.2
|
|
| chr19_-_40186328 | 0.38 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr16_-_9802449 | 0.38 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr3_+_29476085 | 0.38 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr20_-_38746889 | 0.38 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr7_+_8370076 | 0.37 |
ENSDART00000057103
|
zgc:172143
|
zgc:172143 |
| chr2_-_43635777 | 0.37 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr20_+_2039518 | 0.37 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr6_-_38930726 | 0.37 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr12_-_10381282 | 0.37 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr16_-_42872571 | 0.37 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr4_-_5831522 | 0.37 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr1_+_57187794 | 0.36 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr8_+_22955478 | 0.36 |
ENSDART00000099911
|
zgc:136605
|
zgc:136605 |
| chr4_-_15603511 | 0.36 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr15_+_38308421 | 0.35 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr23_+_46183410 | 0.35 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr13_-_1130096 | 0.35 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr1_-_39859626 | 0.35 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr3_-_34032019 | 0.35 |
ENSDART00000151779
|
ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr16_+_52847079 | 0.35 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr17_-_6641535 | 0.35 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr4_+_76637548 | 0.35 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr23_-_27701361 | 0.35 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr13_-_21672131 | 0.35 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr1_+_51615506 | 0.34 |
ENSDART00000152767
|
zgc:165656
|
zgc:165656 |
| chr12_+_22560067 | 0.34 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr11_-_21404358 | 0.34 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr10_+_44956660 | 0.34 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr16_-_45288599 | 0.34 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr20_+_320415 | 0.34 |
ENSDART00000152344
|
si:dkey-119m7.8
|
si:dkey-119m7.8 |
| chr10_+_38610741 | 0.33 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr24_-_12770357 | 0.33 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr1_-_56556326 | 0.33 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr7_+_20344222 | 0.33 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr1_+_23261857 | 0.33 |
ENSDART00000192774
|
CR759875.5
|
|
| chr7_+_22792132 | 0.33 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr7_+_1579510 | 0.33 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr16_+_21426524 | 0.33 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr10_+_31809226 | 0.32 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr21_+_45502621 | 0.32 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr7_-_6425549 | 0.32 |
ENSDART00000173323
|
zgc:165555
|
zgc:165555 |
| chr5_+_27432958 | 0.32 |
ENSDART00000124705
|
histh1l
|
histone H1 like |
| chr23_+_37444933 | 0.32 |
ENSDART00000074385
|
AL954146.1
|
|
| chr23_+_43750646 | 0.32 |
ENSDART00000150047
|
si:ch73-285p12.4
|
si:ch73-285p12.4 |
| chr2_+_38039857 | 0.31 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr7_+_19600262 | 0.31 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr3_+_26135502 | 0.31 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr6_-_28117995 | 0.31 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr20_-_39735952 | 0.31 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr4_-_12790886 | 0.31 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr6_+_33931095 | 0.31 |
ENSDART00000191909
|
orc1
|
origin recognition complex, subunit 1 |
| chr11_-_10643091 | 0.31 |
ENSDART00000192314
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr8_-_14209852 | 0.30 |
ENSDART00000005359
|
def6a
|
differentially expressed in FDCP 6a homolog (mouse) |
| chr4_-_20181964 | 0.30 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr23_-_13875252 | 0.30 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr8_+_22955262 | 0.30 |
ENSDART00000193806
|
zgc:136605
|
zgc:136605 |
| chr21_-_28523548 | 0.30 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr23_+_16807209 | 0.30 |
ENSDART00000141966
|
zgc:114081
|
zgc:114081 |
| chr24_+_20372497 | 0.30 |
ENSDART00000132340
ENSDART00000139989 ENSDART00000168751 ENSDART00000192909 |
plcd1a
|
phospholipase C, delta 1a |
| chr9_-_23891102 | 0.29 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr15_+_19838458 | 0.29 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr21_+_8533533 | 0.29 |
ENSDART00000077924
|
FO834888.1
|
|
| chr19_-_19871211 | 0.29 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 0.8 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 0.7 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.2 | 0.6 | GO:0046222 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.2 | 0.8 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 0.5 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.6 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.5 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 1.3 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) myoblast migration(GO:0051451) |
| 0.1 | 0.3 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.1 | 1.6 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.3 | GO:0014819 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of skeletal muscle contraction(GO:0014819) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.4 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.3 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.6 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 1.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.8 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0072078 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 2.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.2 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.7 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.5 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.8 | GO:0072376 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 0.2 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine monomethylation(GO:0018026) peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:1902765 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.5 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0031936 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.9 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.3 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.4 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.5 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.2 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.8 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 1.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.4 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 1.6 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.8 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.1 | 0.4 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.2 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 1.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.0 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 2.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 1.0 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |