Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
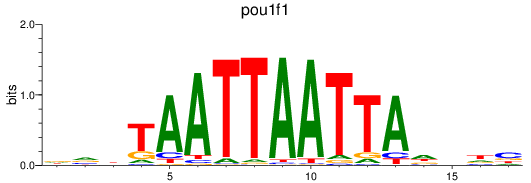
Results for pou1f1
Z-value: 0.74
Transcription factors associated with pou1f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou1f1
|
ENSDARG00000058924 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000110816 | POU class 1 homeobox 1 |
|
pou1f1
|
ENSDARG00000115109 | POU class 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou1f1 | dr11_v1_chr9_-_19161982_19161982 | 0.21 | 7.3e-01 | Click! |
Activity profile of pou1f1 motif
Sorted Z-values of pou1f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17712393 | 0.99 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr21_+_25236297 | 0.55 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr20_-_26822522 | 0.46 |
ENSDART00000146326
ENSDART00000046764 ENSDART00000103234 ENSDART00000143267 |
gmds
|
GDP-mannose 4,6-dehydratase |
| chr22_-_24818066 | 0.41 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr3_+_28939759 | 0.40 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr5_-_63302944 | 0.38 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr7_+_24573721 | 0.35 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr3_+_26145013 | 0.33 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr22_-_24738188 | 0.31 |
ENSDART00000050238
|
vtg1
|
vitellogenin 1 |
| chr25_+_14087045 | 0.29 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr25_-_21031007 | 0.29 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr15_-_33965440 | 0.29 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr6_+_39905021 | 0.28 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr10_-_33621739 | 0.27 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr24_-_26328721 | 0.27 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr10_+_17776981 | 0.26 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr3_+_30922947 | 0.26 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr5_-_44829719 | 0.25 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr2_-_7666021 | 0.25 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr14_+_901847 | 0.24 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr2_+_36004381 | 0.24 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr19_-_5769728 | 0.23 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr10_+_2742499 | 0.23 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr13_+_24756486 | 0.23 |
ENSDART00000137074
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr10_+_38417512 | 0.23 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr17_+_24851951 | 0.22 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr23_+_36460239 | 0.22 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr19_-_5769553 | 0.21 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr23_+_26026383 | 0.21 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr7_+_26029672 | 0.19 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr2_+_36007449 | 0.19 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr8_+_31716872 | 0.19 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr6_-_35046735 | 0.19 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_-_19368435 | 0.18 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr8_-_13574764 | 0.18 |
ENSDART00000076561
|
B3GNT3 (1 of many)
|
si:ch211-126g16.10 |
| chr21_-_44081540 | 0.18 |
ENSDART00000130833
|
FO704810.1
|
|
| chr1_+_58242498 | 0.18 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr8_+_31717175 | 0.18 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr1_-_8963484 | 0.18 |
ENSDART00000188035
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr17_+_30591287 | 0.18 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr14_-_24081929 | 0.18 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr9_-_40765868 | 0.18 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr3_+_36424055 | 0.18 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr3_+_26342768 | 0.18 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr13_-_2010191 | 0.18 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr7_-_66868543 | 0.18 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr25_-_4146947 | 0.18 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr14_+_22022441 | 0.17 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr13_-_37620091 | 0.17 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr18_+_7073130 | 0.17 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr15_-_46779934 | 0.17 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr5_-_66397688 | 0.16 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr20_+_11731039 | 0.15 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr22_-_10156581 | 0.15 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr7_-_54320088 | 0.15 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr19_+_24872159 | 0.15 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr5_+_2815021 | 0.14 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr14_-_4145594 | 0.14 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr19_+_10592778 | 0.14 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr2_+_42330828 | 0.14 |
ENSDART00000134802
ENSDART00000193783 |
ftr09
|
finTRIM family, member 9 |
| chr23_-_16485190 | 0.14 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr5_+_66433287 | 0.14 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr16_-_21810668 | 0.13 |
ENSDART00000156575
|
dicp3.3
|
diverse immunoglobulin domain-containing protein 3.3 |
| chr19_+_43297546 | 0.13 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr20_+_30610547 | 0.13 |
ENSDART00000122256
|
ccr6a
|
chemokine (C-C motif) receptor 6a |
| chr5_-_65000312 | 0.13 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr16_+_54209504 | 0.13 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr12_+_19205086 | 0.13 |
ENSDART00000186573
|
sh3bp1
|
SH3-domain binding protein 1 |
| chr7_-_22632518 | 0.13 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr9_+_21826724 | 0.13 |
ENSDART00000101985
|
zgc:162944
|
zgc:162944 |
| chr21_+_11776328 | 0.13 |
ENSDART00000134469
ENSDART00000081646 |
glrx
|
glutaredoxin (thioltransferase) |
| chr7_+_4694924 | 0.13 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr17_+_45405821 | 0.13 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr13_-_31622195 | 0.12 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr2_+_9990491 | 0.12 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr2_-_10877765 | 0.12 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr9_-_25989989 | 0.12 |
ENSDART00000090052
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr5_+_71802014 | 0.12 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr23_-_3511630 | 0.12 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr22_+_1556948 | 0.12 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr9_-_52920305 | 0.11 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr7_+_4694762 | 0.11 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr20_+_25225112 | 0.11 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr19_+_12915498 | 0.11 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr23_+_19813677 | 0.11 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr14_-_17588345 | 0.11 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr23_-_6522099 | 0.11 |
ENSDART00000092214
ENSDART00000183380 ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr17_-_27048537 | 0.11 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr4_+_70151160 | 0.11 |
ENSDART00000111816
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr17_-_32413147 | 0.11 |
ENSDART00000149102
|
grhl1
|
grainyhead-like transcription factor 1 |
| chr12_-_48188928 | 0.11 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr25_+_4751879 | 0.11 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr15_+_34592215 | 0.10 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr2_+_36112273 | 0.10 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr19_+_43780970 | 0.10 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr9_+_54290896 | 0.10 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr25_+_22320738 | 0.10 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_+_36560019 | 0.10 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr11_-_29737088 | 0.10 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr5_+_6672870 | 0.10 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr23_+_32101202 | 0.10 |
ENSDART00000000992
|
zgc:56699
|
zgc:56699 |
| chr23_-_16484383 | 0.10 |
ENSDART00000187839
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr4_+_12966640 | 0.09 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr22_+_508290 | 0.09 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr22_-_5529241 | 0.09 |
ENSDART00000157893
|
CABZ01064968.1
|
|
| chr12_-_28363111 | 0.09 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr6_+_49551614 | 0.09 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr4_+_70015788 | 0.09 |
ENSDART00000161133
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr11_-_10456387 | 0.09 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr1_+_5402476 | 0.09 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr23_-_1660708 | 0.09 |
ENSDART00000175138
|
CU693481.1
|
|
| chr4_-_948776 | 0.09 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr14_+_32430982 | 0.09 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr2_+_20605925 | 0.09 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr22_+_29990448 | 0.09 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr6_-_24053404 | 0.09 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr11_-_10456553 | 0.08 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr2_+_16160906 | 0.08 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr1_-_58975098 | 0.08 |
ENSDART00000189899
|
CABZ01114581.1
|
|
| chr6_+_12527725 | 0.08 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr8_-_50888806 | 0.08 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr7_-_22632690 | 0.08 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr22_+_18469004 | 0.08 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr24_+_26329018 | 0.08 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr11_+_14321113 | 0.08 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr22_-_22337382 | 0.08 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr14_+_1170968 | 0.08 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr3_+_23092762 | 0.08 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr8_+_52637507 | 0.08 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr23_-_23401305 | 0.07 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr5_+_21891305 | 0.07 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr4_-_73136420 | 0.07 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr2_-_9059955 | 0.07 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr7_+_20383841 | 0.07 |
ENSDART00000052906
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr18_+_13275735 | 0.07 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr9_+_2574122 | 0.07 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr20_+_7584211 | 0.07 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr6_+_612594 | 0.07 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr8_+_2487250 | 0.07 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr11_-_1509773 | 0.07 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr22_+_2751887 | 0.07 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr10_+_13000669 | 0.07 |
ENSDART00000158919
ENSDART00000172625 |
lpar1
|
lysophosphatidic acid receptor 1 |
| chr23_-_29394505 | 0.07 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr8_+_2487883 | 0.07 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr3_+_23029934 | 0.07 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr7_-_67248829 | 0.07 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr2_-_59205393 | 0.07 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr23_+_41679586 | 0.06 |
ENSDART00000067662
|
CU914487.1
|
|
| chr4_-_58883559 | 0.06 |
ENSDART00000164546
|
si:dkey-28i19.4
|
si:dkey-28i19.4 |
| chr5_+_12528693 | 0.06 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr9_+_38588081 | 0.06 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr25_+_5972690 | 0.06 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr8_+_28259347 | 0.06 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr1_+_52792439 | 0.06 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr14_-_1355544 | 0.06 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr6_+_9427641 | 0.06 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr25_+_3217419 | 0.06 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr16_+_46401756 | 0.06 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr16_-_27566552 | 0.06 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr25_-_18739924 | 0.06 |
ENSDART00000156328
|
si:dkeyp-93a5.2
|
si:dkeyp-93a5.2 |
| chr16_-_38118003 | 0.06 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr24_+_23840821 | 0.06 |
ENSDART00000128595
ENSDART00000127188 |
pola1
|
polymerase (DNA directed), alpha 1 |
| chr19_-_32150078 | 0.06 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr7_+_49800478 | 0.06 |
ENSDART00000174254
|
PAMR1 (1 of many)
|
si:ch211-102l7.3 |
| chr5_-_61445697 | 0.06 |
ENSDART00000050898
|
ncf1
|
neutrophil cytosolic factor 1 |
| chr12_+_39203745 | 0.05 |
ENSDART00000153661
|
si:dkeyp-106c3.2
|
si:dkeyp-106c3.2 |
| chr5_+_12888260 | 0.05 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr20_-_40755614 | 0.05 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr14_+_16287968 | 0.05 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr3_+_3681116 | 0.05 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr10_-_45210184 | 0.05 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr6_+_40714811 | 0.05 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr9_+_48761455 | 0.05 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr21_-_1644414 | 0.05 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr1_+_51721851 | 0.05 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr2_-_24061575 | 0.05 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr13_+_9432501 | 0.05 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr11_+_44617021 | 0.05 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr1_-_58002973 | 0.05 |
ENSDART00000140390
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr23_+_4709607 | 0.05 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr1_+_44127292 | 0.05 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr5_-_1203455 | 0.05 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr16_+_46401576 | 0.05 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr7_+_4702269 | 0.05 |
ENSDART00000139880
|
si:ch211-225k7.4
|
si:ch211-225k7.4 |
| chr1_-_55118745 | 0.05 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr1_-_54063520 | 0.05 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr8_-_13046089 | 0.05 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr11_-_2838699 | 0.05 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr2_+_2169337 | 0.04 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr23_+_33991509 | 0.04 |
ENSDART00000145161
|
fam50a
|
family with sequence similarity 50, member A |
| chr19_+_1688727 | 0.04 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr15_-_43284021 | 0.04 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr8_+_50953776 | 0.04 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr13_+_18321140 | 0.04 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr17_-_12336987 | 0.04 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr14_-_30971264 | 0.04 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr8_+_10001805 | 0.04 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr7_+_4657987 | 0.04 |
ENSDART00000144321
ENSDART00000142344 |
si:dkey-83f18.2
|
si:dkey-83f18.2 |
| chr2_+_49799470 | 0.04 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr22_+_5532003 | 0.04 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou1f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.1 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.9 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.1 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0002020 | protease binding(GO:0002020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |