Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
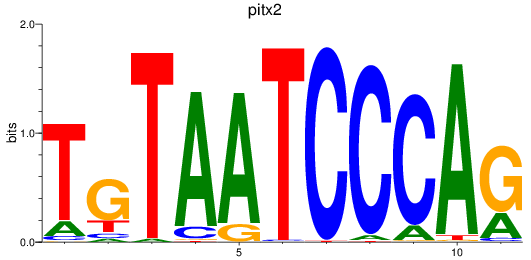
Results for pitx2
Z-value: 1.54
Transcription factors associated with pitx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx2
|
ENSDARG00000036194 | paired-like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx2 | dr11_v1_chr14_+_36223097_36223097 | 0.49 | 4.0e-01 | Click! |
Activity profile of pitx2 motif
Sorted Z-values of pitx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_45510977 | 3.16 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr23_+_28731379 | 1.14 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr7_-_48173440 | 1.13 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr5_-_23277939 | 1.12 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr11_-_4235811 | 1.01 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr4_+_6643421 | 0.96 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr13_-_30028103 | 0.94 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr11_+_25101220 | 0.90 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr8_+_41533268 | 0.87 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr2_-_2020044 | 0.86 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr12_+_47917971 | 0.79 |
ENSDART00000185933
|
tbata
|
thymus, brain and testes associated |
| chr19_-_9829965 | 0.77 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr25_-_13839743 | 0.76 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr21_+_22630297 | 0.75 |
ENSDART00000147175
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr19_-_25271155 | 0.74 |
ENSDART00000104027
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr19_+_12762887 | 0.74 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr25_+_21833287 | 0.71 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_+_26075230 | 0.70 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr23_-_24856025 | 0.68 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr21_+_22630627 | 0.67 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr4_+_8168514 | 0.67 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr7_-_57933736 | 0.65 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr21_+_13366353 | 0.64 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr23_+_22267374 | 0.64 |
ENSDART00000079035
|
rap1gap
|
RAP1 GTPase activating protein |
| chr11_-_44409856 | 0.63 |
ENSDART00000162886
|
il1rapl1b
|
interleukin 1 receptor accessory protein-like 1b |
| chr18_+_26337869 | 0.60 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr15_-_12319065 | 0.59 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr25_+_21832938 | 0.57 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr25_+_6306885 | 0.57 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr9_-_29321625 | 0.56 |
ENSDART00000158689
ENSDART00000014047 ENSDART00000122602 |
pth2r
|
parathyroid hormone 2 receptor |
| chr14_-_2270973 | 0.56 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr10_-_43392267 | 0.55 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr7_-_22132265 | 0.54 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr9_+_22631672 | 0.54 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr2_-_42871286 | 0.53 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr22_+_696931 | 0.52 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr9_-_12034444 | 0.51 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr16_-_13004166 | 0.51 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr19_+_37701450 | 0.51 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr24_+_24461341 | 0.50 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr5_-_29488245 | 0.49 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr5_-_65158203 | 0.49 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr13_-_30027730 | 0.49 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr5_+_52067723 | 0.48 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr21_+_41743493 | 0.48 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr6_-_46162585 | 0.46 |
ENSDART00000017309
|
ca16b
|
carbonic anhydrase XVI b |
| chr11_-_18468570 | 0.46 |
ENSDART00000155474
ENSDART00000193869 |
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr4_-_9579299 | 0.45 |
ENSDART00000183079
ENSDART00000192968 ENSDART00000091809 |
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr10_+_29963518 | 0.45 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr6_-_9581949 | 0.45 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr21_+_3854414 | 0.44 |
ENSDART00000122699
|
miga2
|
mitoguardin 2 |
| chr2_-_2096055 | 0.44 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr23_+_8989168 | 0.43 |
ENSDART00000034380
|
xkr7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr12_+_48390715 | 0.43 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr1_-_1894722 | 0.43 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr18_-_40508528 | 0.41 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr13_-_24448278 | 0.41 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr14_+_44545092 | 0.41 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr13_+_16522608 | 0.40 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr5_-_38094130 | 0.40 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr20_+_41640687 | 0.40 |
ENSDART00000138686
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_+_40604951 | 0.40 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr22_+_17509422 | 0.40 |
ENSDART00000088419
|
march2
|
membrane-associated ring finger (C3HC4) 2 |
| chr8_+_14886452 | 0.39 |
ENSDART00000146589
|
soat1
|
sterol O-acyltransferase 1 |
| chr22_-_23625351 | 0.39 |
ENSDART00000192588
ENSDART00000163423 |
cfhl4
|
complement factor H like 4 |
| chr9_+_22632126 | 0.38 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr2_+_6253246 | 0.38 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr8_+_7737062 | 0.38 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr1_+_33383644 | 0.38 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr6_-_12135741 | 0.38 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr14_+_8504410 | 0.38 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr16_-_43971258 | 0.37 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr23_+_40109353 | 0.37 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr18_-_39702327 | 0.37 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr25_-_7999756 | 0.36 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr14_-_2206476 | 0.36 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr23_-_15330168 | 0.36 |
ENSDART00000035865
ENSDART00000143635 |
sulf2b
|
sulfatase 2b |
| chr21_-_32467099 | 0.36 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr5_+_31049742 | 0.36 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr21_+_20715020 | 0.36 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr20_+_18225329 | 0.36 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr10_+_15777258 | 0.36 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr5_+_62052538 | 0.35 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr6_+_21740672 | 0.35 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr10_+_15777064 | 0.34 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr1_+_38758261 | 0.34 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr2_-_17947389 | 0.34 |
ENSDART00000190089
ENSDART00000191872 ENSDART00000184039 ENSDART00000179791 |
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr23_-_24825863 | 0.34 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr10_-_7857494 | 0.33 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr9_-_32753535 | 0.33 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr2_-_6292510 | 0.33 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr12_+_27068525 | 0.33 |
ENSDART00000188634
|
srcap
|
Snf2-related CREBBP activator protein |
| chr3_+_26019426 | 0.33 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr2_-_32738535 | 0.33 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr8_+_50983551 | 0.33 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr17_+_33495194 | 0.32 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr17_+_5915875 | 0.32 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr17_+_25289431 | 0.32 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr14_+_30730749 | 0.32 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr10_+_37173029 | 0.31 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr5_-_71765780 | 0.31 |
ENSDART00000011955
|
gpsm1b
|
G protein signaling modulator 1b |
| chr13_+_15816573 | 0.31 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr8_+_694218 | 0.31 |
ENSDART00000147753
|
rnf165b
|
ring finger protein 165b |
| chr1_+_38758445 | 0.31 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr7_+_38255418 | 0.30 |
ENSDART00000052354
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr20_-_18731268 | 0.30 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr18_-_40509006 | 0.30 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr11_-_43002262 | 0.30 |
ENSDART00000172477
ENSDART00000181513 |
CABZ01069998.1
|
|
| chr21_+_10756154 | 0.30 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr25_+_26844028 | 0.29 |
ENSDART00000127274
ENSDART00000156179 |
sema7a
|
semaphorin 7A |
| chr10_-_7821686 | 0.29 |
ENSDART00000121531
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr9_+_34127005 | 0.29 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr6_-_36182115 | 0.29 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr11_-_27057572 | 0.28 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr7_+_30970045 | 0.28 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr7_+_28901786 | 0.28 |
ENSDART00000113998
|
dok4
|
docking protein 4 |
| chr7_-_19369002 | 0.28 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr10_+_37182626 | 0.28 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr25_-_19090479 | 0.27 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr25_+_10458990 | 0.27 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr10_+_21511495 | 0.27 |
ENSDART00000178395
|
BX957322.3
|
|
| chr13_-_49666615 | 0.27 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr15_-_20024205 | 0.27 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr8_-_54304381 | 0.27 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr11_+_6456146 | 0.26 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr5_-_20205075 | 0.26 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr17_-_16342388 | 0.26 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr11_-_29563437 | 0.25 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr19_-_40776267 | 0.25 |
ENSDART00000189038
|
calcr
|
calcitonin receptor |
| chr5_+_33301005 | 0.25 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr15_-_21014270 | 0.24 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr23_-_45682136 | 0.24 |
ENSDART00000164646
|
fam160a1b
|
family with sequence similarity 160, member A1b |
| chr1_+_7546259 | 0.24 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr18_+_36806545 | 0.24 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr15_-_21014015 | 0.24 |
ENSDART00000144991
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr18_-_21806613 | 0.24 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr19_+_42898239 | 0.24 |
ENSDART00000051724
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr2_+_31475772 | 0.24 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr23_-_28239750 | 0.23 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr25_-_13703826 | 0.23 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr18_+_48608366 | 0.23 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr9_-_53537989 | 0.23 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr2_+_49522178 | 0.23 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr17_-_24714837 | 0.22 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr5_+_483965 | 0.22 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr2_+_2967255 | 0.22 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr13_+_9896845 | 0.21 |
ENSDART00000169076
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr12_+_17436904 | 0.21 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr5_-_72125551 | 0.21 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr12_+_30653047 | 0.21 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr4_-_20232974 | 0.21 |
ENSDART00000193353
|
stk38l
|
serine/threonine kinase 38 like |
| chr23_+_39413163 | 0.21 |
ENSDART00000184254
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr2_-_11258547 | 0.20 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr24_+_7336807 | 0.20 |
ENSDART00000137010
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr21_+_10866421 | 0.20 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr7_+_29167744 | 0.20 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr23_+_21638258 | 0.20 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr3_-_37351225 | 0.20 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr19_+_4142529 | 0.20 |
ENSDART00000162913
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr18_+_507835 | 0.20 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr12_+_30706158 | 0.19 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr17_+_21760032 | 0.19 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr10_+_26926654 | 0.19 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr21_-_39177564 | 0.19 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr15_+_11693624 | 0.19 |
ENSDART00000193630
ENSDART00000161930 |
strn4
|
striatin, calmodulin binding protein 4 |
| chr5_+_64840656 | 0.19 |
ENSDART00000073953
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr13_-_30149973 | 0.19 |
ENSDART00000041515
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr8_+_16990120 | 0.18 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr23_-_31400792 | 0.18 |
ENSDART00000132736
|
lca5
|
Leber congenital amaurosis 5 |
| chr20_+_36233873 | 0.18 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr9_+_14028157 | 0.18 |
ENSDART00000134603
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr18_+_507618 | 0.18 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr3_+_52806347 | 0.18 |
ENSDART00000168151
ENSDART00000098826 |
lmx1al
|
LIM homeobox transcription factor 1, alpha-like |
| chr10_+_8534750 | 0.17 |
ENSDART00000183960
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr6_-_6248893 | 0.17 |
ENSDART00000124662
|
rtn4a
|
reticulon 4a |
| chr14_-_2004291 | 0.17 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr9_+_307863 | 0.17 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr23_+_45282858 | 0.17 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr18_-_17001056 | 0.17 |
ENSDART00000112627
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr1_+_33322555 | 0.17 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr19_+_10661520 | 0.17 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr18_-_17000842 | 0.17 |
ENSDART00000079832
|
tph2
|
tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) |
| chr20_-_25436389 | 0.17 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr5_-_68456987 | 0.17 |
ENSDART00000157810
ENSDART00000170819 |
ephb4a
|
eph receptor B4a |
| chr9_+_23748133 | 0.17 |
ENSDART00000180758
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr8_+_17184602 | 0.16 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr5_+_37978501 | 0.16 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr19_-_3193912 | 0.16 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr14_-_2264494 | 0.16 |
ENSDART00000191149
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr19_-_31802296 | 0.16 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr11_-_26832685 | 0.16 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr18_+_6054816 | 0.16 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr17_+_28624321 | 0.16 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr16_-_45910050 | 0.15 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr4_+_49664256 | 0.15 |
ENSDART00000155251
|
znf974
|
zinc finger protein 974 |
| chr12_+_28995942 | 0.15 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr17_-_43552894 | 0.15 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr13_+_50151407 | 0.15 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr7_+_47316944 | 0.15 |
ENSDART00000173534
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr6_-_39903393 | 0.15 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 3.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.4 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.6 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 1.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.6 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 1.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.3 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.6 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.3 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 1.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.5 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 1.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.3 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.4 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.5 | GO:0033135 | regulation of peptidyl-serine phosphorylation(GO:0033135) |
| 0.0 | 1.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.6 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.8 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.1 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.6 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 3.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.4 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.7 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |