Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
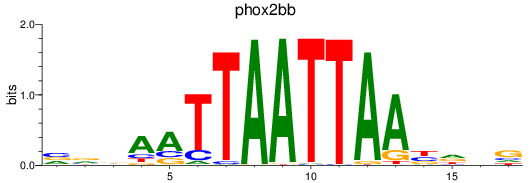
Results for phox2bb
Z-value: 0.41
Transcription factors associated with phox2bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
phox2bb
|
ENSDARG00000091029 | paired like homeobox 2Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| phox2bb | dr11_v1_chr14_-_17068712_17068712 | 1.00 | 3.2e-05 | Click! |
Activity profile of phox2bb motif
Sorted Z-values of phox2bb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_50608099 | 0.62 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr16_-_12173399 | 0.52 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr16_-_12173554 | 0.47 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr10_-_27049170 | 0.46 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr17_+_15433518 | 0.43 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 0.43 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr14_-_4556896 | 0.41 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr6_-_11768198 | 0.40 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr23_+_16638639 | 0.33 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr9_-_43538328 | 0.32 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr16_+_46111849 | 0.30 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr21_-_13123176 | 0.30 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr9_+_54039006 | 0.26 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr4_-_1801519 | 0.25 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr17_-_21418340 | 0.24 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr2_+_2223837 | 0.23 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr3_-_48716422 | 0.22 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr9_-_37749973 | 0.22 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr5_+_29851433 | 0.22 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr15_-_14552101 | 0.19 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_+_27513859 | 0.18 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr14_-_470505 | 0.18 |
ENSDART00000067147
|
ANKRD50
|
ankyrin repeat domain 50 |
| chr25_-_13490744 | 0.17 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr6_+_11249706 | 0.17 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_11250033 | 0.14 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr2_-_16216568 | 0.14 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_15797663 | 0.14 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr21_-_22635245 | 0.13 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr4_-_2219705 | 0.12 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr15_-_22074315 | 0.12 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr9_-_32177117 | 0.12 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr1_+_10318089 | 0.10 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr6_+_11250316 | 0.10 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr24_+_28953089 | 0.09 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr17_+_31914877 | 0.08 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr17_+_22577472 | 0.07 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr17_-_4245902 | 0.07 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr3_-_26787430 | 0.07 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr5_+_60590796 | 0.06 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr22_+_2751887 | 0.06 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr7_+_38811800 | 0.06 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr8_-_44926077 | 0.05 |
ENSDART00000084417
|
timm17b
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr5_+_63302660 | 0.05 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr17_-_16422654 | 0.04 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr11_-_2838699 | 0.03 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr23_-_2901167 | 0.02 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr2_+_38373272 | 0.02 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr9_-_27398369 | 0.01 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr1_-_31534089 | 0.01 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr22_+_23359369 | 0.01 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr2_-_17393216 | 0.01 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr1_+_21731382 | 0.01 |
ENSDART00000054395
|
pax5
|
paired box 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of phox2bb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |