Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
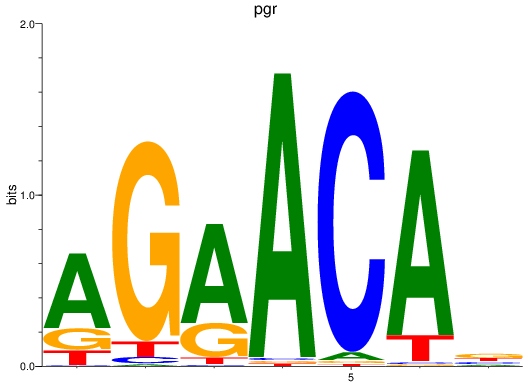
Results for pgr
Z-value: 1.52
Transcription factors associated with pgr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pgr
|
ENSDARG00000035966 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pgr | dr11_v1_chr18_+_41820039_41820039 | 0.15 | 8.1e-01 | Click! |
Activity profile of pgr motif
Sorted Z-values of pgr motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_10866421 | 1.14 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr9_-_22831836 | 1.06 |
ENSDART00000142585
|
neb
|
nebulin |
| chr6_+_41096058 | 0.79 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr21_+_20901505 | 0.76 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr1_+_7538909 | 0.75 |
ENSDART00000148276
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr5_-_32308526 | 0.74 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr7_+_39389273 | 0.72 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr22_-_278328 | 0.70 |
ENSDART00000098072
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr4_-_14328997 | 0.68 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr16_+_47308856 | 0.67 |
ENSDART00000170103
|
col28a1b
|
collagen, type XXVIII, alpha 1b |
| chr6_+_23887314 | 0.58 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr16_+_23924040 | 0.55 |
ENSDART00000161124
|
si:dkey-7f3.14
|
si:dkey-7f3.14 |
| chr11_-_1024483 | 0.54 |
ENSDART00000017551
|
slc6a1b
|
solute carrier family 6 (neurotransmitter transporter), member 1b |
| chr17_-_6730247 | 0.53 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr3_+_49397115 | 0.50 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr3_+_16612574 | 0.48 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr25_+_31277415 | 0.46 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr7_+_25033924 | 0.45 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr3_+_16449490 | 0.45 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr2_+_47581997 | 0.44 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr21_-_22737228 | 0.44 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr19_-_42556086 | 0.43 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr2_+_47582488 | 0.43 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr11_-_33919014 | 0.41 |
ENSDART00000181203
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr20_+_572037 | 0.41 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr16_-_2414063 | 0.40 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr6_+_41099787 | 0.40 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr15_-_19677511 | 0.40 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr7_-_66864756 | 0.40 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr23_-_21471022 | 0.40 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr7_+_25036188 | 0.39 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr6_-_6254432 | 0.39 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr13_-_31622195 | 0.39 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr13_+_35745572 | 0.35 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr1_-_10914523 | 0.35 |
ENSDART00000007013
|
dmd
|
dystrophin |
| chr1_+_56229674 | 0.35 |
ENSDART00000127714
|
c3a.2
|
complement component c3a, duplicate 2 |
| chr2_-_37893910 | 0.34 |
ENSDART00000143027
|
hbl2
|
hexose-binding lectin 2 |
| chr2_+_47582681 | 0.34 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr7_+_35245607 | 0.34 |
ENSDART00000193422
ENSDART00000173888 |
amfrb
|
autocrine motility factor receptor b |
| chr23_-_45501177 | 0.33 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr18_+_5547185 | 0.33 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr3_+_34220194 | 0.33 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr22_-_24880824 | 0.33 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr15_-_32383529 | 0.33 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr9_-_34944604 | 0.33 |
ENSDART00000140563
ENSDART00000136812 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr6_+_27339962 | 0.32 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr6_+_60112200 | 0.31 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr16_-_6821927 | 0.31 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr7_-_51953613 | 0.31 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr19_-_20113696 | 0.30 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr18_-_39200557 | 0.30 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr9_-_43001898 | 0.29 |
ENSDART00000138515
|
ttn.2
|
titin, tandem duplicate 2 |
| chr15_+_43093044 | 0.29 |
ENSDART00000141125
|
kcne4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr2_-_51757328 | 0.28 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr15_-_32383340 | 0.28 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr24_+_3307857 | 0.28 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr2_-_24996441 | 0.28 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr9_-_41277347 | 0.28 |
ENSDART00000181213
|
glsb
|
glutaminase b |
| chr11_+_35364445 | 0.27 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr21_+_25643880 | 0.27 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr9_+_34127005 | 0.26 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr22_-_23668356 | 0.26 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr18_+_38908903 | 0.26 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr20_+_51466046 | 0.25 |
ENSDART00000178065
|
tlr5b
|
toll-like receptor 5b |
| chr23_-_21453614 | 0.25 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr9_-_8454060 | 0.25 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr19_-_9867001 | 0.25 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr6_-_60031693 | 0.24 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr6_-_20707846 | 0.24 |
ENSDART00000169836
|
col18a1b
|
collagen type XVIII alpha 1 chain b |
| chr20_+_30445971 | 0.24 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr22_+_18477934 | 0.23 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr20_+_33904258 | 0.23 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr21_+_28478663 | 0.23 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr19_+_2279051 | 0.22 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr11_+_21076872 | 0.22 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr8_-_14050758 | 0.22 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr6_-_49078263 | 0.22 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr3_-_58650057 | 0.22 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr7_+_52761841 | 0.22 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr7_+_73780936 | 0.22 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr17_+_45454943 | 0.22 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr20_+_43942278 | 0.21 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr1_+_51615672 | 0.21 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr15_+_37197494 | 0.21 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr14_-_2050057 | 0.21 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr3_-_3448095 | 0.21 |
ENSDART00000078886
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr21_+_35327589 | 0.21 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr23_+_21455152 | 0.20 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr9_-_46842179 | 0.20 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr17_-_465285 | 0.20 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr4_-_17823424 | 0.20 |
ENSDART00000024822
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr7_+_30823749 | 0.20 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr17_+_20504196 | 0.19 |
ENSDART00000190539
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr25_+_4541211 | 0.19 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr18_-_46063773 | 0.19 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr11_+_13223625 | 0.19 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr3_+_32691275 | 0.19 |
ENSDART00000191838
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr7_+_34297271 | 0.19 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr21_+_18353703 | 0.19 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr15_+_46605482 | 0.18 |
ENSDART00000184449
|
CABZ01044048.1
|
|
| chr19_-_42651615 | 0.18 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr19_-_29437108 | 0.18 |
ENSDART00000052108
ENSDART00000074497 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr10_-_22249444 | 0.18 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr3_-_28048475 | 0.18 |
ENSDART00000150888
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_+_66634167 | 0.18 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr24_-_38816725 | 0.18 |
ENSDART00000063231
|
nog2
|
noggin 2 |
| chr11_+_2202987 | 0.18 |
ENSDART00000190008
ENSDART00000173139 |
hoxc6b
|
homeobox C6b |
| chr6_+_54248705 | 0.17 |
ENSDART00000162469
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr10_+_21786656 | 0.17 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr24_+_23742690 | 0.17 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr13_+_38430466 | 0.17 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr22_+_12595144 | 0.17 |
ENSDART00000140054
ENSDART00000060979 ENSDART00000139826 |
zgc:92335
|
zgc:92335 |
| chr16_-_29458806 | 0.17 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr15_+_32710300 | 0.17 |
ENSDART00000167857
|
postnb
|
periostin, osteoblast specific factor b |
| chr1_-_48922273 | 0.17 |
ENSDART00000150254
|
si:ch211-112b1.2
|
si:ch211-112b1.2 |
| chr7_-_44672095 | 0.17 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr11_+_13224281 | 0.17 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr1_+_33969015 | 0.17 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr21_-_10446405 | 0.17 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr9_+_31752628 | 0.16 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr23_+_21459263 | 0.16 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr12_+_33151246 | 0.16 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr16_-_12496632 | 0.16 |
ENSDART00000019941
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr4_-_4706893 | 0.16 |
ENSDART00000093005
|
CABZ01020835.1
|
|
| chr3_-_5664123 | 0.16 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr13_-_34683370 | 0.16 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr5_-_28029558 | 0.16 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr22_-_20575679 | 0.16 |
ENSDART00000089033
|
lingo3a
|
leucine rich repeat and Ig domain containing 3a |
| chr16_-_41936932 | 0.16 |
ENSDART00000111956
|
gramd1a
|
GRAM domain containing 1A |
| chr6_-_1865323 | 0.16 |
ENSDART00000155775
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr15_-_24869826 | 0.16 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr4_-_5302162 | 0.15 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr25_+_3564220 | 0.15 |
ENSDART00000170005
|
zgc:194202
|
zgc:194202 |
| chr6_-_39764995 | 0.15 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr2_+_50608099 | 0.15 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr22_-_8006342 | 0.15 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr12_+_13405445 | 0.15 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr15_-_35406564 | 0.15 |
ENSDART00000086051
|
mecom
|
MDS1 and EVI1 complex locus |
| chr6_-_13308813 | 0.15 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr25_-_210730 | 0.15 |
ENSDART00000187580
|
FP236318.1
|
|
| chr18_-_16179129 | 0.14 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr23_-_30781510 | 0.14 |
ENSDART00000183364
|
myt1a
|
myelin transcription factor 1a |
| chr3_-_2589704 | 0.14 |
ENSDART00000187239
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr17_-_26507289 | 0.14 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr6_+_8622228 | 0.14 |
ENSDART00000110470
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr12_+_24561832 | 0.14 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr2_-_36932253 | 0.14 |
ENSDART00000124217
|
map1sb
|
microtubule-associated protein 1Sb |
| chr6_-_40098641 | 0.14 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr3_-_42981739 | 0.14 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr20_-_26846028 | 0.14 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr6_+_2951645 | 0.14 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr6_+_12462079 | 0.14 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr1_-_39976492 | 0.14 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr24_+_32592748 | 0.14 |
ENSDART00000188256
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr6_-_39765546 | 0.13 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr1_+_57311901 | 0.13 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr13_-_25719628 | 0.13 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr15_+_15316326 | 0.13 |
ENSDART00000125864
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr18_-_6634424 | 0.13 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr15_+_32710094 | 0.13 |
ENSDART00000159592
|
postnb
|
periostin, osteoblast specific factor b |
| chr15_+_17343319 | 0.13 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr23_-_44677315 | 0.13 |
ENSDART00000143054
|
kif1c
|
kinesin family member 1C |
| chr16_+_23921777 | 0.13 |
ENSDART00000163213
|
apoa4b.3
|
apolipoprotein A-IV b, tandem duplicate 3 |
| chr12_+_15165736 | 0.13 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr12_+_11352630 | 0.13 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr14_-_2209742 | 0.12 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr9_-_42871756 | 0.12 |
ENSDART00000191396
|
ttn.1
|
titin, tandem duplicate 1 |
| chr15_-_36352983 | 0.12 |
ENSDART00000059786
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr20_+_25911342 | 0.12 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr23_-_30781875 | 0.12 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr18_+_29145681 | 0.12 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr1_+_57757456 | 0.12 |
ENSDART00000152650
|
si:dkey-1c7.1
|
si:dkey-1c7.1 |
| chr4_+_26628822 | 0.12 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr12_+_13404784 | 0.12 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr15_+_37950556 | 0.12 |
ENSDART00000153947
|
si:dkey-238d18.12
|
si:dkey-238d18.12 |
| chr2_+_27932781 | 0.12 |
ENSDART00000019555
|
mc4r
|
melanocortin 4 receptor |
| chr7_+_61764040 | 0.12 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr21_+_26620867 | 0.12 |
ENSDART00000176008
|
esrra
|
estrogen-related receptor alpha |
| chr2_-_31018062 | 0.12 |
ENSDART00000140841
ENSDART00000087026 |
emilin2a
|
elastin microfibril interfacer 2a |
| chr4_+_74551606 | 0.12 |
ENSDART00000174339
|
kcna1b
|
potassium voltage-gated channel, shaker-related subfamily, member 1b |
| chr2_+_25868404 | 0.12 |
ENSDART00000143517
ENSDART00000187743 |
slc7a14a
|
solute carrier family 7, member 14a |
| chr5_-_67911111 | 0.12 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr5_+_37966505 | 0.12 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr2_-_32352946 | 0.12 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr7_+_4194055 | 0.12 |
ENSDART00000105073
|
si:dkey-28d5.14
|
si:dkey-28d5.14 |
| chr2_+_55916911 | 0.12 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr3_+_42497814 | 0.11 |
ENSDART00000184653
|
BX537305.1
|
|
| chr19_+_25478203 | 0.11 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr9_+_44832032 | 0.11 |
ENSDART00000002633
|
frzb
|
frizzled related protein |
| chr17_+_15041647 | 0.11 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr20_+_9785952 | 0.11 |
ENSDART00000152403
|
ddhd1b
|
DDHD domain containing 1b |
| chr14_-_2189889 | 0.11 |
ENSDART00000181557
ENSDART00000106707 |
pcdh2ab9
pcdh2ab11
|
protocadherin 2 alpha b 9 protocadherin 2 alpha b 11 |
| chr1_+_9290103 | 0.11 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr24_-_22702017 | 0.11 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr9_+_4306122 | 0.11 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr4_+_10616626 | 0.11 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr15_+_28482862 | 0.11 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr21_-_25601648 | 0.11 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr20_+_33532296 | 0.11 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr2_-_38992304 | 0.11 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pgr
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.4 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.3 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 0.3 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.2 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 1.7 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 0.3 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.3 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.1 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.2 | GO:0050996 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.2 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0006145 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.5 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0071376 | neutrophil homeostasis(GO:0001780) response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.0 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 0.0 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.0 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.0 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.4 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.1 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |