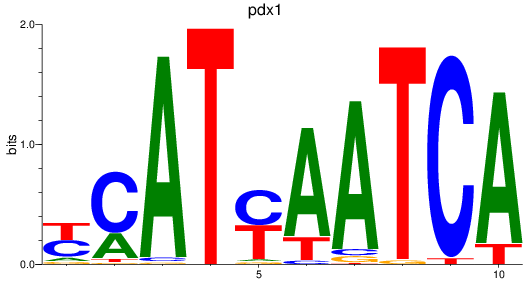
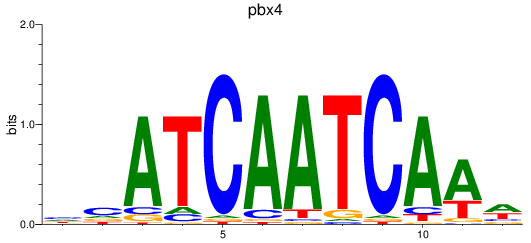
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for pdx1_pbx4
Z-value: 1.84
Transcription factors associated with pdx1_pbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pdx1
|
ENSDARG00000002779 | pancreatic and duodenal homeobox 1 |
|
pdx1
|
ENSDARG00000109275 | pancreatic and duodenal homeobox 1 |
|
pbx4
|
ENSDARG00000052150 | pre-B-cell leukemia transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pbx4 | dr11_v1_chr3_+_53052769_53052769 | -0.07 | 9.1e-01 | Click! |
| pdx1 | dr11_v1_chr24_+_21684189_21684189 | -0.07 | 9.1e-01 | Click! |
Activity profile of pdx1_pbx4 motif
Sorted Z-values of pdx1_pbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 17.54 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr21_-_23308286 | 1.68 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr2_+_47623202 | 1.64 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr24_-_6898302 | 1.35 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr21_-_23307653 | 1.25 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr15_+_4632782 | 1.17 |
ENSDART00000156012
|
si:dkey-35i13.1
|
si:dkey-35i13.1 |
| chr24_-_6897884 | 1.16 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr20_-_8443425 | 1.06 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr5_-_23280098 | 1.03 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr4_+_21129752 | 1.00 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr13_-_24447332 | 0.95 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr10_-_39011514 | 0.78 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr12_+_39685485 | 0.77 |
ENSDART00000163403
|
LO017650.1
|
|
| chr3_-_32818607 | 0.76 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr24_-_4973765 | 0.72 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr10_-_17745345 | 0.70 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr19_-_26869103 | 0.69 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr11_-_1509773 | 0.68 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr7_+_29952719 | 0.68 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr15_-_10341048 | 0.66 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr19_+_37925616 | 0.64 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr9_-_22834860 | 0.63 |
ENSDART00000146486
|
neb
|
nebulin |
| chr2_-_54039293 | 0.59 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr3_+_33300522 | 0.58 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr4_-_25485404 | 0.58 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_-_9277986 | 0.57 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr21_-_43079161 | 0.56 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr9_-_23217196 | 0.55 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr1_+_17676745 | 0.55 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr9_+_42066030 | 0.52 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr18_-_36135799 | 0.52 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr3_-_16068236 | 0.48 |
ENSDART00000157315
|
si:dkey-81l17.6
|
si:dkey-81l17.6 |
| chr24_-_40009446 | 0.47 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr18_+_783936 | 0.46 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr3_-_62380146 | 0.45 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr4_-_5291256 | 0.45 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr22_+_3223489 | 0.45 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr24_+_34113424 | 0.42 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr20_-_29420713 | 0.42 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr17_+_27434626 | 0.41 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr23_-_11870962 | 0.41 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr4_-_4780667 | 0.41 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr23_+_6077503 | 0.41 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr8_+_22931427 | 0.41 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr11_-_13341483 | 0.41 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr14_+_35237613 | 0.40 |
ENSDART00000163465
|
ebf3a
|
early B cell factor 3a |
| chr19_-_10395683 | 0.40 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr13_+_38990939 | 0.39 |
ENSDART00000145979
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr18_+_41495841 | 0.39 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr11_-_13341051 | 0.39 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr14_+_36231126 | 0.39 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr15_-_33925851 | 0.38 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr4_-_4261673 | 0.38 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr25_-_4482449 | 0.38 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr18_-_38088099 | 0.37 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr15_-_27710513 | 0.37 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr22_-_38508112 | 0.37 |
ENSDART00000192004
|
klc4
|
kinesin light chain 4 |
| chr20_+_15015557 | 0.36 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr16_-_13004166 | 0.36 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr24_-_24881996 | 0.36 |
ENSDART00000145712
|
crhb
|
corticotropin releasing hormone b |
| chr17_-_200316 | 0.36 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr14_-_31893996 | 0.35 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr19_-_10243148 | 0.35 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr8_+_49778756 | 0.35 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr7_-_52842007 | 0.35 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr22_+_24715282 | 0.35 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr17_-_12385308 | 0.35 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr1_-_8428736 | 0.34 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr16_-_44649053 | 0.34 |
ENSDART00000184807
|
CR925804.2
|
|
| chr4_-_27301356 | 0.34 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr16_+_26777473 | 0.34 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr25_+_15354095 | 0.34 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr21_+_31151197 | 0.33 |
ENSDART00000137728
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr13_+_14006118 | 0.33 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr25_-_29363934 | 0.33 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr5_-_29643930 | 0.33 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr8_+_49778486 | 0.33 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr1_-_19215336 | 0.33 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr4_+_842010 | 0.32 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr25_-_13842618 | 0.32 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr12_+_38770654 | 0.32 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr22_+_15438513 | 0.31 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr13_+_38302665 | 0.31 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr21_-_26918901 | 0.31 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr1_-_7917062 | 0.31 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr4_+_13810811 | 0.30 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr18_-_42172101 | 0.30 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr8_-_34052019 | 0.29 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr5_-_7834582 | 0.29 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr16_-_18960333 | 0.29 |
ENSDART00000143185
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr17_+_23937262 | 0.29 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr20_-_42702832 | 0.29 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr2_+_24199276 | 0.29 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr14_-_32403554 | 0.29 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr24_+_39186940 | 0.28 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr2_+_27010439 | 0.28 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr21_+_25181003 | 0.28 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr12_-_26406323 | 0.28 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr7_+_50109239 | 0.28 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr24_+_41915878 | 0.28 |
ENSDART00000171523
|
TMEM200C
|
transmembrane protein 200C |
| chr25_+_21833287 | 0.28 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr14_-_2199573 | 0.28 |
ENSDART00000124485
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr24_+_37370064 | 0.28 |
ENSDART00000185870
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr17_+_19626479 | 0.28 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr13_+_24842857 | 0.28 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr20_-_29418620 | 0.28 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr7_+_4384863 | 0.27 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr23_+_40460333 | 0.27 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr1_-_40227166 | 0.27 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr13_+_30421472 | 0.27 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr13_+_23282549 | 0.26 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr1_-_681116 | 0.26 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr11_+_25129013 | 0.26 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr5_-_50992690 | 0.26 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr17_+_50127648 | 0.26 |
ENSDART00000156460
|
galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr18_+_13792490 | 0.25 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr15_-_25527580 | 0.25 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr6_-_53143667 | 0.25 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr2_+_42724404 | 0.25 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr7_+_4386753 | 0.25 |
ENSDART00000172915
|
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr11_-_4220167 | 0.24 |
ENSDART00000185406
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr8_+_23165749 | 0.24 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr20_+_522457 | 0.24 |
ENSDART00000138585
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr19_-_25149034 | 0.24 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr2_+_24199073 | 0.24 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr20_+_34717403 | 0.24 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr3_-_12890670 | 0.23 |
ENSDART00000159934
ENSDART00000188607 |
btbd17b
|
BTB (POZ) domain containing 17b |
| chr3_-_59981476 | 0.23 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_-_26023678 | 0.23 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr16_+_30301539 | 0.23 |
ENSDART00000186018
|
LO017848.1
|
|
| chr7_+_30875273 | 0.23 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr15_+_30917282 | 0.22 |
ENSDART00000129474
|
omgb
|
oligodendrocyte myelin glycoprotein b |
| chr5_+_23622177 | 0.22 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr24_+_32472155 | 0.22 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr7_+_26629084 | 0.22 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr2_-_10188598 | 0.22 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr5_-_34185115 | 0.22 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr10_-_26744131 | 0.22 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr13_+_22249636 | 0.22 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr15_+_43093044 | 0.22 |
ENSDART00000141125
|
kcne4
|
potassium voltage-gated channel, Isk-related family, member 4 |
| chr18_+_28102620 | 0.22 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr6_-_53144336 | 0.22 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr7_+_46252993 | 0.21 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr21_-_20328375 | 0.21 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr4_+_26496489 | 0.21 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_+_56366395 | 0.21 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr10_-_27741793 | 0.21 |
ENSDART00000129369
ENSDART00000192440 ENSDART00000189808 ENSDART00000138149 |
auts2a
si:dkey-33o22.1
|
autism susceptibility candidate 2a si:dkey-33o22.1 |
| chr10_-_20445549 | 0.21 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr5_+_44654535 | 0.21 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr17_-_6399920 | 0.21 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr7_-_19940473 | 0.20 |
ENSDART00000127669
|
prox1b
|
prospero homeobox 1b |
| chr25_-_16818978 | 0.20 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr23_+_23182037 | 0.20 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr18_-_25051846 | 0.20 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr24_+_27548474 | 0.20 |
ENSDART00000105774
ENSDART00000123368 |
ek1
|
eph-like kinase 1 |
| chr6_+_39232245 | 0.20 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr22_-_33679277 | 0.20 |
ENSDART00000169948
|
FO904977.1
|
|
| chr5_+_64842730 | 0.20 |
ENSDART00000144732
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr18_-_14836600 | 0.19 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr20_-_54381034 | 0.19 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr25_-_7925269 | 0.19 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr12_-_31009315 | 0.19 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr19_-_10425140 | 0.19 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr23_+_216012 | 0.19 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr7_-_41964877 | 0.19 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr5_-_11573490 | 0.19 |
ENSDART00000109577
|
FO704871.1
|
|
| chr25_+_20215964 | 0.19 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr3_-_16227490 | 0.19 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr19_+_9790806 | 0.18 |
ENSDART00000155948
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr6_-_47246948 | 0.18 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr9_-_23156908 | 0.18 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr23_+_27703749 | 0.18 |
ENSDART00000027224
|
lmbr1l
|
limb development membrane protein 1-like |
| chr1_-_23557877 | 0.18 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr25_-_7925019 | 0.18 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr1_+_39387876 | 0.18 |
ENSDART00000084891
ENSDART00000148701 |
tenm3
|
teneurin transmembrane protein 3 |
| chr13_-_36545258 | 0.18 |
ENSDART00000186171
|
FP103009.1
|
|
| chr24_+_18948665 | 0.18 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr12_+_34827808 | 0.18 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr16_-_29458806 | 0.18 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr17_+_31914877 | 0.18 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr21_+_20715020 | 0.18 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr19_-_867071 | 0.18 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr6_+_18367388 | 0.18 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr8_-_19051906 | 0.18 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr2_-_10192459 | 0.18 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr3_-_28857195 | 0.17 |
ENSDART00000114319
|
si:ch211-76l23.7
|
si:ch211-76l23.7 |
| chr15_-_34213898 | 0.17 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr2_+_35240764 | 0.17 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr16_+_14707960 | 0.17 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr16_+_23282655 | 0.17 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr22_-_3595439 | 0.17 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr6_+_4872883 | 0.17 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr8_-_31393429 | 0.17 |
ENSDART00000192369
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr15_-_5467477 | 0.17 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr20_+_46897504 | 0.17 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr12_+_9703172 | 0.17 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr2_+_2223837 | 0.16 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr21_+_43669943 | 0.16 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr10_+_26571174 | 0.16 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr13_-_16191674 | 0.16 |
ENSDART00000147868
|
vwc2
|
von Willebrand factor C domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pdx1_pbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 2.9 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 1.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.6 | GO:0051503 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.6 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.6 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.6 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.2 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 17.1 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 0.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.5 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.1 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.6 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.6 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.6 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.3 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.9 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.0 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0048644 | muscle organ morphogenesis(GO:0048644) |
| 0.0 | 0.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.3 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0016199 | axon choice point recognition(GO:0016198) axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.0 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 2.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 1.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 2.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 2.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0070699 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |