Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
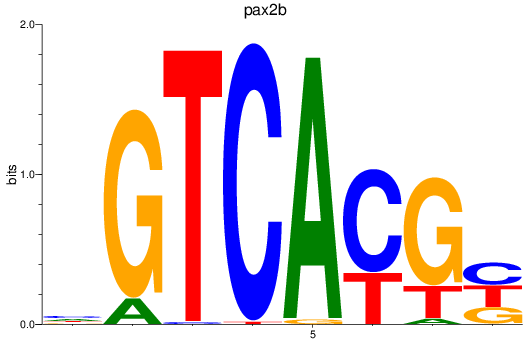
Results for pax2b
Z-value: 1.81
Transcription factors associated with pax2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2b
|
ENSDARG00000032578 | paired box 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2b | dr11_v1_chr12_-_45876387_45876387 | 0.83 | 8.0e-02 | Click! |
Activity profile of pax2b motif
Sorted Z-values of pax2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_25112269 | 1.40 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr24_-_7632187 | 1.26 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr16_-_45069882 | 1.21 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr22_-_11136625 | 1.10 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr23_-_4915118 | 1.03 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr15_-_163586 | 1.02 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr15_+_28989573 | 1.02 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| chr24_+_39158283 | 1.02 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr16_+_10777116 | 0.96 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr24_-_6158933 | 0.78 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr1_+_10720294 | 0.77 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr24_-_17023392 | 0.77 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr13_-_23007813 | 0.77 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr4_+_9669717 | 0.75 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr11_-_10798021 | 0.73 |
ENSDART00000167112
ENSDART00000179725 ENSDART00000091923 ENSDART00000185825 |
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr3_-_15264698 | 0.73 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr23_-_24343363 | 0.73 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr12_+_29054907 | 0.73 |
ENSDART00000152936
|
gabrz
|
gamma-aminobutyric acid (GABA) A receptor, zeta |
| chr2_-_31302615 | 0.71 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr7_+_38770167 | 0.68 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr20_+_20637866 | 0.68 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr21_-_39628771 | 0.68 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr6_-_12135741 | 0.68 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr8_-_40327397 | 0.66 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr20_+_27393668 | 0.66 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr2_-_24996441 | 0.66 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr24_+_31334209 | 0.65 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr10_+_32683089 | 0.64 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr13_+_421231 | 0.63 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr16_-_2558653 | 0.63 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr2_-_38000276 | 0.62 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr8_+_50983551 | 0.61 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr21_-_45613564 | 0.61 |
ENSDART00000160324
|
LO018363.1
|
|
| chr10_+_15777258 | 0.61 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr7_-_42206720 | 0.60 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr21_+_13861589 | 0.60 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr1_-_38815361 | 0.59 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr16_+_31853919 | 0.59 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr7_-_41812015 | 0.59 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr16_+_36906693 | 0.59 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr11_+_37275448 | 0.58 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr3_+_43460696 | 0.57 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr20_+_29743904 | 0.57 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr3_+_40576447 | 0.56 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr19_-_20114149 | 0.56 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr16_+_30117798 | 0.56 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr7_-_41812355 | 0.56 |
ENSDART00000016105
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr21_-_42100471 | 0.55 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr24_-_33756003 | 0.55 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr19_-_11546516 | 0.55 |
ENSDART00000103646
|
kcng2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr4_-_5302866 | 0.55 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr2_+_30786773 | 0.55 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr2_-_30460293 | 0.55 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr14_+_22172047 | 0.54 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr10_+_35953068 | 0.54 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr12_-_33359052 | 0.53 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_21062706 | 0.53 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr15_+_22311803 | 0.53 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr4_-_240737 | 0.53 |
ENSDART00000166186
|
RERG
|
si:cabz01085950.1 |
| chr1_-_46924801 | 0.53 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr18_+_26337869 | 0.52 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr18_+_2837563 | 0.52 |
ENSDART00000171495
ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr10_-_6775271 | 0.52 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr9_+_219124 | 0.52 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr2_+_42177113 | 0.52 |
ENSDART00000056441
|
tmeff1a
|
transmembrane protein with EGF-like and two follistatin-like domains 1a |
| chr3_-_29968015 | 0.52 |
ENSDART00000077119
ENSDART00000139310 |
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr19_-_30510930 | 0.52 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr24_-_31846366 | 0.51 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr8_+_25761654 | 0.51 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr7_-_30082931 | 0.51 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr13_+_23843712 | 0.51 |
ENSDART00000057611
|
oprm1
|
opioid receptor, mu 1 |
| chr19_-_103289 | 0.51 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr2_-_44283554 | 0.51 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr12_+_24342303 | 0.51 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr15_-_20956384 | 0.51 |
ENSDART00000135770
|
tbcela
|
tubulin folding cofactor E-like a |
| chr20_+_50956369 | 0.50 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr12_-_33972798 | 0.50 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr5_+_57320113 | 0.50 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr22_-_21676364 | 0.49 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr2_-_54387550 | 0.49 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr2_+_30787128 | 0.48 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr12_-_29624638 | 0.48 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr17_-_20143946 | 0.48 |
ENSDART00000138911
|
actn2b
|
actinin, alpha 2b |
| chr23_+_44741500 | 0.48 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr23_-_12345764 | 0.48 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr8_+_24854600 | 0.48 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr5_+_33301005 | 0.48 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr21_+_22400951 | 0.48 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr2_-_51096647 | 0.48 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr9_-_31747106 | 0.47 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr15_-_23342752 | 0.47 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr20_-_20610812 | 0.47 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr13_+_12739283 | 0.47 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr14_-_44790817 | 0.46 |
ENSDART00000098640
|
grhpra
|
glyoxylate reductase/hydroxypyruvate reductase a |
| chr10_-_7974155 | 0.46 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr10_+_23060391 | 0.46 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr6_-_10780698 | 0.46 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr23_-_29505463 | 0.46 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr17_-_22062364 | 0.46 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr20_-_45423498 | 0.45 |
ENSDART00000098424
|
trib2
|
tribbles pseudokinase 2 |
| chr2_+_50626476 | 0.45 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr17_-_20711735 | 0.44 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr11_-_1509773 | 0.44 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr2_+_301898 | 0.44 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr15_-_19250543 | 0.43 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr4_-_16124417 | 0.43 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr8_+_31248917 | 0.43 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr6_-_51386656 | 0.43 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_+_16633951 | 0.43 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr4_-_16836006 | 0.42 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr3_-_50277959 | 0.42 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr6_+_59808677 | 0.42 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr25_+_33939728 | 0.42 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr16_+_32995882 | 0.42 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr23_-_29505645 | 0.42 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr14_-_24410673 | 0.42 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr15_+_16897554 | 0.42 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr2_+_48288461 | 0.42 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr25_-_19443421 | 0.42 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr7_+_48999723 | 0.41 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr18_+_39074139 | 0.41 |
ENSDART00000142390
|
gnb5b
|
guanine nucleotide binding protein (G protein), beta 5b |
| chr13_+_23669659 | 0.41 |
ENSDART00000188518
ENSDART00000078079 ENSDART00000134973 |
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr17_-_7861219 | 0.41 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr18_+_43183749 | 0.41 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr7_+_42206543 | 0.41 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr11_-_1291012 | 0.41 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr15_-_47115787 | 0.40 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr8_-_6943155 | 0.40 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr2_-_42871286 | 0.40 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr9_-_28103097 | 0.40 |
ENSDART00000146284
|
kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr21_+_13205859 | 0.40 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr3_-_34337969 | 0.40 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr3_+_15271943 | 0.40 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr15_+_28685892 | 0.39 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr9_-_1703761 | 0.39 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr2_-_32486080 | 0.39 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr3_-_22829710 | 0.39 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr6_-_30839763 | 0.39 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr23_-_36724575 | 0.39 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr18_+_17428506 | 0.39 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr13_+_35925490 | 0.39 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr6_+_27667359 | 0.39 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr12_-_48960308 | 0.39 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr10_+_26571174 | 0.38 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr10_-_2527342 | 0.38 |
ENSDART00000184168
|
CU856539.1
|
|
| chr10_+_25219728 | 0.38 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr3_-_35554809 | 0.38 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr5_-_23179319 | 0.38 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr8_+_29742237 | 0.38 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr4_-_5247335 | 0.38 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr10_+_20108557 | 0.38 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr19_-_5103313 | 0.38 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_+_3449780 | 0.38 |
ENSDART00000163849
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr15_-_1534232 | 0.37 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr2_+_20430366 | 0.37 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr15_+_28685625 | 0.37 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr19_-_32888758 | 0.37 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr12_-_15620090 | 0.37 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr17_+_5895500 | 0.37 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr11_+_37137196 | 0.37 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr7_-_41013575 | 0.37 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr1_-_40086978 | 0.37 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr19_-_29437108 | 0.37 |
ENSDART00000052108
ENSDART00000074497 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr8_+_53452681 | 0.37 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr4_-_77267787 | 0.36 |
ENSDART00000190346
|
zgc:174310
|
zgc:174310 |
| chr9_+_19421841 | 0.36 |
ENSDART00000159090
ENSDART00000084771 |
pde9a
|
phosphodiesterase 9A |
| chr15_+_16387088 | 0.36 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr20_+_40766645 | 0.36 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr1_-_10473630 | 0.36 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr2_+_16846772 | 0.36 |
ENSDART00000183564
ENSDART00000126718 |
fam131a
|
family with sequence similarity 131, member A |
| chr13_-_9841806 | 0.36 |
ENSDART00000101949
|
sfxn4
|
sideroflexin 4 |
| chr2_-_6292510 | 0.36 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr14_+_8947282 | 0.36 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_+_8183393 | 0.36 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr25_-_8916913 | 0.36 |
ENSDART00000104629
ENSDART00000131748 |
furinb
|
furin (paired basic amino acid cleaving enzyme) b |
| chr14_-_48033073 | 0.36 |
ENSDART00000193115
ENSDART00000169300 ENSDART00000188036 ENSDART00000183432 ENSDART00000180973 |
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr20_-_40766387 | 0.36 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr2_+_42871831 | 0.36 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr10_-_35220285 | 0.35 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr23_-_16692312 | 0.35 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr3_-_21106093 | 0.35 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr20_-_45040916 | 0.35 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr15_-_12500938 | 0.35 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr25_+_13791627 | 0.35 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr9_+_43797902 | 0.35 |
ENSDART00000020550
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr9_-_43375205 | 0.35 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr7_+_67486807 | 0.35 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr25_+_35706493 | 0.35 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr8_+_36582728 | 0.35 |
ENSDART00000049230
|
pqbp1
|
polyglutamine binding protein 1 |
| chr6_+_48041759 | 0.34 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr9_-_44583167 | 0.34 |
ENSDART00000140689
ENSDART00000191842 |
pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr13_-_33022372 | 0.34 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr25_+_1591964 | 0.34 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr24_-_10828560 | 0.34 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr19_+_33093577 | 0.34 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr17_+_890988 | 0.34 |
ENSDART00000186843
|
CABZ01078858.1
|
|
| chr9_-_18742704 | 0.34 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr8_+_14792830 | 0.33 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr16_+_21242491 | 0.33 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr1_+_52929185 | 0.33 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 0.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.5 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 0.5 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.7 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 0.5 | GO:0015824 | proline transport(GO:0015824) |
| 0.2 | 0.6 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.4 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.5 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.5 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.5 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.5 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 0.4 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.4 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 2.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.0 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.5 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.7 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.6 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.3 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.4 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.3 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.3 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.3 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.3 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.3 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.8 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0090133 | coronary vasculature development(GO:0060976) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 3.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.2 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.2 | GO:0021856 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.1 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:0043268 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 1.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.2 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.4 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.1 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:1902946 | protein localization to endosome(GO:0036010) protein localization to early endosome(GO:1902946) |
| 0.0 | 1.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.3 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.5 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.4 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.2 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 1.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 1.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 2.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.5 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 0.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 0.7 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.2 | 0.5 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 0.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.5 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.8 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 3.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.3 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 0.3 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.3 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 1.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 1.0 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.3 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.2 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 1.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 1.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.6 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.0 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.4 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 1.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |