Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
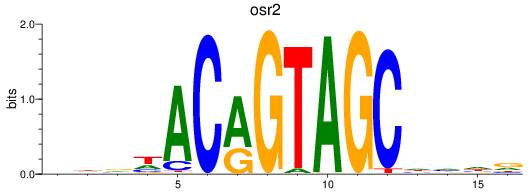
Results for osr2
Z-value: 0.61
Transcription factors associated with osr2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr2
|
ENSDARG00000038006 | odd-skipped related transciption factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr2 | dr11_v1_chr16_-_54405976_54405976 | 0.74 | 1.6e-01 | Click! |
Activity profile of osr2 motif
Sorted Z-values of osr2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_29418620 | 0.34 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr7_+_42206543 | 0.26 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr22_-_28871787 | 0.22 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr14_+_36246726 | 0.21 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr15_-_39848822 | 0.20 |
ENSDART00000155230
|
si:dkey-263j23.1
|
si:dkey-263j23.1 |
| chr14_+_23717165 | 0.19 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr13_+_30696286 | 0.18 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr18_-_27316599 | 0.18 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr3_-_28120092 | 0.18 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr19_-_25519310 | 0.17 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr5_+_13373593 | 0.17 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr20_+_30490682 | 0.16 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr6_+_46431848 | 0.15 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr19_-_25519612 | 0.15 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr7_+_42206847 | 0.15 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr6_+_54576520 | 0.15 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr23_+_36095260 | 0.14 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr7_-_42206720 | 0.13 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr12_-_38548299 | 0.13 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr5_-_48307804 | 0.12 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr19_-_6134802 | 0.11 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr8_+_10835456 | 0.11 |
ENSDART00000151388
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr18_-_39200557 | 0.11 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr3_+_25907266 | 0.10 |
ENSDART00000170324
ENSDART00000192633 |
tom1
|
target of myb1 membrane trafficking protein |
| chr10_+_38512270 | 0.10 |
ENSDART00000109752
|
serpinh1a
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1a |
| chr16_-_13680692 | 0.10 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr14_+_48964628 | 0.09 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr12_+_28856151 | 0.09 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr6_-_40884453 | 0.09 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr21_-_25250594 | 0.07 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr7_+_38090515 | 0.07 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr3_-_32958505 | 0.06 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr19_-_9503473 | 0.06 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr13_+_29778610 | 0.06 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr18_-_46010 | 0.05 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_+_35344031 | 0.04 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr11_+_31285127 | 0.03 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr18_-_45736 | 0.03 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr23_-_36003282 | 0.02 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr25_-_19395476 | 0.02 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr13_-_27621246 | 0.02 |
ENSDART00000102232
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr8_-_22639794 | 0.02 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr2_+_5446882 | 0.02 |
ENSDART00000083260
|
dusp28
|
dual specificity phosphatase 28 |
| chr2_+_3986083 | 0.02 |
ENSDART00000188979
|
mkxb
|
mohawk homeobox b |
| chr16_-_13612650 | 0.01 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr10_-_13239367 | 0.01 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |