Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
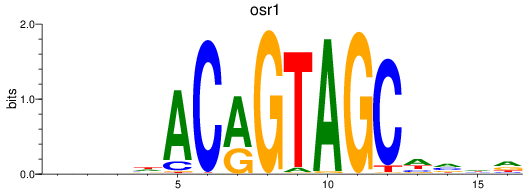
Results for osr1
Z-value: 1.13
Transcription factors associated with osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
osr1
|
ENSDARG00000014091 | odd-skipped related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| osr1 | dr11_v1_chr13_+_32148338_32148338 | 0.48 | 4.1e-01 | Click! |
Activity profile of osr1 motif
Sorted Z-values of osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_45601103 | 1.03 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr21_+_45627775 | 0.98 |
ENSDART00000185466
|
irf1b
|
interferon regulatory factor 1b |
| chr3_-_32958505 | 0.94 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr7_+_22767678 | 0.84 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr16_-_31469065 | 0.82 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr9_-_33063083 | 0.82 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr19_-_25519310 | 0.78 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr5_+_13373593 | 0.77 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr13_+_30696286 | 0.75 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr3_-_16784280 | 0.74 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr6_+_54576520 | 0.71 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr4_-_8040436 | 0.71 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr5_+_29831235 | 0.70 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr22_+_661711 | 0.69 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr16_-_46660680 | 0.67 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr14_+_4151379 | 0.67 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr19_-_5769553 | 0.66 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr14_+_29769336 | 0.64 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr21_+_4702773 | 0.61 |
ENSDART00000147404
|
si:dkey-102g19.3
|
si:dkey-102g19.3 |
| chr1_-_59130695 | 0.60 |
ENSDART00000152560
|
FP015850.1
|
|
| chr19_-_25519612 | 0.59 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr5_-_55981288 | 0.58 |
ENSDART00000146616
|
si:dkey-189h5.6
|
si:dkey-189h5.6 |
| chr21_-_25295087 | 0.58 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr22_+_29991834 | 0.56 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr1_+_54650048 | 0.55 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr7_+_29955368 | 0.54 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr6_+_52947186 | 0.53 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr7_-_8315179 | 0.51 |
ENSDART00000184049
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr8_-_36469117 | 0.50 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr2_-_36918709 | 0.49 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr9_-_5263947 | 0.47 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr6_+_52947699 | 0.47 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr18_-_977075 | 0.46 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr7_+_29954709 | 0.45 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr5_-_30978381 | 0.44 |
ENSDART00000127787
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr3_-_61181018 | 0.44 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr20_+_53521648 | 0.43 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr14_+_21905492 | 0.41 |
ENSDART00000054437
|
tbc1d10c
|
TBC1 domain family, member 10C |
| chr1_+_40199074 | 0.41 |
ENSDART00000179679
ENSDART00000136849 |
si:ch211-113e8.5
|
si:ch211-113e8.5 |
| chr13_+_12456412 | 0.41 |
ENSDART00000057762
|
dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr25_-_31433512 | 0.41 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr16_-_21810668 | 0.41 |
ENSDART00000156575
|
dicp3.3
|
diverse immunoglobulin domain-containing protein 3.3 |
| chr20_+_16881883 | 0.40 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr7_-_17635717 | 0.40 |
ENSDART00000080704
|
nitr6b
|
novel immune-type receptor 6b |
| chr15_-_952891 | 0.39 |
ENSDART00000153780
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr11_-_40457325 | 0.39 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr17_-_6198823 | 0.38 |
ENSDART00000028407
ENSDART00000193636 |
ptk2ba
|
protein tyrosine kinase 2 beta, a |
| chr21_-_7940043 | 0.38 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr21_-_11657043 | 0.38 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr20_+_53522059 | 0.38 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr21_-_26490186 | 0.38 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr4_-_1824836 | 0.37 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr11_-_33960318 | 0.36 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr19_+_31576849 | 0.36 |
ENSDART00000134107
ENSDART00000088401 |
acot13
|
acyl-CoA thioesterase 13 |
| chr6_+_19950107 | 0.36 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr2_+_36114194 | 0.36 |
ENSDART00000113547
|
traj39
|
T-cell receptor alpha joining 39 |
| chr1_+_55643198 | 0.35 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr14_-_1538600 | 0.35 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr14_+_51098036 | 0.35 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr8_-_2506327 | 0.34 |
ENSDART00000101125
ENSDART00000125124 |
rpl6
|
ribosomal protein L6 |
| chr13_-_45523026 | 0.34 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr7_+_38090515 | 0.34 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr21_-_32036597 | 0.34 |
ENSDART00000114964
|
zgc:165573
|
zgc:165573 |
| chr1_-_58064738 | 0.33 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr3_-_29899172 | 0.33 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr8_+_4798158 | 0.33 |
ENSDART00000031650
|
hsp70l
|
heat shock cognate 70-kd protein, like |
| chr8_+_21297217 | 0.33 |
ENSDART00000142836
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr15_-_576135 | 0.32 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr2_-_15318786 | 0.32 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr2_-_4868002 | 0.32 |
ENSDART00000189835
|
tfr1a
|
transferrin receptor 1a |
| chr25_+_37435720 | 0.31 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr7_+_64936903 | 0.31 |
ENSDART00000114592
|
adgrg3
|
adhesion G protein-coupled receptor G3 |
| chr9_+_33423170 | 0.30 |
ENSDART00000141542
|
si:dkey-216e24.9
|
si:dkey-216e24.9 |
| chr18_-_45736 | 0.30 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr4_-_3805992 | 0.30 |
ENSDART00000190125
|
si:dkey-61f9.1
|
si:dkey-61f9.1 |
| chr23_+_4709607 | 0.30 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr22_-_5006801 | 0.29 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr5_+_6617401 | 0.29 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr19_-_35361556 | 0.29 |
ENSDART00000012167
|
ndufs5
|
NADH dehydrogenase (ubiquinone) Fe-S protein 5 |
| chr24_+_19518303 | 0.29 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr16_-_29387215 | 0.28 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr1_-_8140763 | 0.28 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr12_-_38548299 | 0.27 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr8_+_48858132 | 0.27 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr5_+_25317061 | 0.27 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr23_+_25232711 | 0.27 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr16_+_46684855 | 0.27 |
ENSDART00000058325
|
lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr4_+_69262318 | 0.26 |
ENSDART00000188841
|
si:ch211-209j12.6
|
si:ch211-209j12.6 |
| chr3_+_31093455 | 0.25 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr21_+_33454147 | 0.25 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr23_+_43809484 | 0.25 |
ENSDART00000149534
|
si:ch211-149b19.3
|
si:ch211-149b19.3 |
| chr16_-_28876479 | 0.25 |
ENSDART00000047862
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr19_+_10794072 | 0.25 |
ENSDART00000151518
ENSDART00000142614 |
lipea
|
lipase, hormone-sensitive a |
| chr13_+_25397098 | 0.25 |
ENSDART00000132953
|
gsto2
|
glutathione S-transferase omega 2 |
| chr8_+_22472584 | 0.25 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr17_-_23631400 | 0.25 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr7_-_71758613 | 0.25 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr16_-_33095161 | 0.24 |
ENSDART00000187648
|
dopey1
|
dopey family member 1 |
| chr23_-_42752550 | 0.24 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr15_-_434503 | 0.24 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr3_+_23063718 | 0.24 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr3_-_48980319 | 0.24 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr5_+_37749503 | 0.24 |
ENSDART00000148586
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr3_-_10751491 | 0.23 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr13_+_25396896 | 0.23 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr3_-_32973511 | 0.23 |
ENSDART00000131808
ENSDART00000103182 |
casp6l2
|
caspase 6, apoptosis-related cysteine peptidase, like 2 |
| chr3_-_2072630 | 0.23 |
ENSDART00000189404
|
BX005442.3
|
|
| chr3_+_41558682 | 0.23 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr12_-_29305533 | 0.23 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr3_-_55139127 | 0.23 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr23_-_30041065 | 0.22 |
ENSDART00000131209
ENSDART00000127192 |
ccdc187
|
coiled-coil domain containing 187 |
| chr23_+_5104743 | 0.22 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr10_+_29840725 | 0.22 |
ENSDART00000127268
|
BX120005.1
|
|
| chr6_-_52566574 | 0.22 |
ENSDART00000098421
|
uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr10_-_22506044 | 0.22 |
ENSDART00000100497
|
zgc:172339
|
zgc:172339 |
| chr8_-_1219815 | 0.22 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr8_-_20245892 | 0.22 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr15_+_404891 | 0.22 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr3_+_32571929 | 0.21 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr7_-_7904683 | 0.21 |
ENSDART00000192822
|
CABZ01030278.1
|
|
| chr7_+_24390939 | 0.21 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr7_-_17600415 | 0.21 |
ENSDART00000080717
|
nitr6b
|
novel immune-type receptor 6b |
| chr2_+_20539402 | 0.21 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr1_+_230363 | 0.21 |
ENSDART00000153285
|
tfdp1b
|
transcription factor Dp-1, b |
| chr3_+_32129632 | 0.21 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr4_+_64034834 | 0.21 |
ENSDART00000160654
|
si:dkey-179k24.5
|
si:dkey-179k24.5 |
| chr13_+_40686133 | 0.21 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr23_+_4324625 | 0.21 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr1_+_44767657 | 0.21 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chr2_-_37896646 | 0.21 |
ENSDART00000075931
|
hbl1
|
hexose-binding lectin 1 |
| chr20_-_26866111 | 0.20 |
ENSDART00000077767
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr17_+_32622933 | 0.20 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr15_+_22390076 | 0.20 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr21_-_15674802 | 0.20 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr24_-_33461313 | 0.20 |
ENSDART00000163706
|
si:ch73-173p19.2
|
si:ch73-173p19.2 |
| chr3_-_26115558 | 0.19 |
ENSDART00000078671
ENSDART00000124762 |
hsp70.1
|
heat shock cognate 70-kd protein, tandem duplicate 1 |
| chr15_+_35933094 | 0.19 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr6_-_11812224 | 0.19 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr16_+_23331633 | 0.19 |
ENSDART00000187455
|
krtcap2
|
keratinocyte associated protein 2 |
| chr7_-_18508815 | 0.19 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr23_-_42752387 | 0.19 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr15_-_5016039 | 0.18 |
ENSDART00000156458
|
defbl3
|
defensin, beta-like 3 |
| chr9_-_33121535 | 0.18 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr18_-_46258612 | 0.18 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr8_+_40284973 | 0.18 |
ENSDART00000018401
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr25_-_7670391 | 0.18 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr2_-_37462462 | 0.18 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr11_-_36350876 | 0.18 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr9_-_34269066 | 0.18 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr13_+_28417297 | 0.18 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr21_-_25250594 | 0.17 |
ENSDART00000163862
|
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr1_+_44826367 | 0.17 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr18_-_27316599 | 0.17 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr2_+_39021282 | 0.17 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr5_-_16274058 | 0.17 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr24_+_1294176 | 0.17 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr3_-_4303262 | 0.17 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr5_-_16472719 | 0.17 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr23_+_7379728 | 0.16 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr14_+_5383060 | 0.16 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr7_+_31871830 | 0.16 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr25_-_7670616 | 0.16 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr22_-_21314821 | 0.16 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr11_+_44617021 | 0.16 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr3_+_6291635 | 0.16 |
ENSDART00000185055
ENSDART00000157707 |
si:ch211-12p12.2
|
si:ch211-12p12.2 |
| chr22_+_19473804 | 0.15 |
ENSDART00000135085
ENSDART00000147673 ENSDART00000156818 |
si:dkey-78l4.7
|
si:dkey-78l4.7 |
| chr25_-_35143360 | 0.15 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr11_-_29996344 | 0.15 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr2_-_43942595 | 0.15 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr14_-_22108718 | 0.15 |
ENSDART00000054410
|
med19a
|
mediator complex subunit 19a |
| chr1_-_18585046 | 0.15 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr7_+_4648398 | 0.14 |
ENSDART00000148099
|
si:dkey-83f18.3
|
si:dkey-83f18.3 |
| chr1_-_34750169 | 0.14 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr17_-_45378473 | 0.14 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr17_-_45104750 | 0.14 |
ENSDART00000075520
|
aldh6a1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_-_9134045 | 0.14 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr8_-_36554675 | 0.14 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr2_+_10821579 | 0.14 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr1_-_31657854 | 0.14 |
ENSDART00000085309
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr23_-_9925568 | 0.14 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr16_+_20871021 | 0.14 |
ENSDART00000006429
|
hibadhb
|
3-hydroxyisobutyrate dehydrogenase b |
| chr4_+_35189063 | 0.14 |
ENSDART00000185091
|
si:dkey-269p2.1
|
si:dkey-269p2.1 |
| chr1_+_17593392 | 0.14 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr7_-_59564011 | 0.13 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr15_-_39848822 | 0.13 |
ENSDART00000155230
|
si:dkey-263j23.1
|
si:dkey-263j23.1 |
| chr20_+_43113465 | 0.13 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
| chr21_-_40834413 | 0.13 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr1_+_26676758 | 0.13 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr23_-_4704938 | 0.12 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr3_-_34040063 | 0.12 |
ENSDART00000151503
|
ighv8-4
|
immunoglobulin heavy variable 8-4 |
| chr12_-_33789218 | 0.12 |
ENSDART00000193258
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr2_-_22363460 | 0.12 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr11_+_31285127 | 0.12 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr7_+_17374560 | 0.12 |
ENSDART00000172272
ENSDART00000073467 |
nitr7a
|
novel immune-type receptor 7a |
| chr10_-_34089779 | 0.12 |
ENSDART00000140070
|
pimr144
|
Pim proto-oncogene, serine/threonine kinase, related 144 |
| chr4_+_43408004 | 0.12 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr5_+_26847190 | 0.12 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr5_-_36949476 | 0.12 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr11_-_36279602 | 0.12 |
ENSDART00000122531
ENSDART00000103064 ENSDART00000125616 |
nfya
|
nuclear transcription factor Y, alpha |
| chr19_+_43341115 | 0.11 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.4 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 1.0 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.3 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.3 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.2 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.1 | 0.2 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.7 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.3 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.4 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.4 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.5 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.0 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.0 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.1 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 1.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.0 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0070513 | death domain binding(GO:0070513) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |