Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
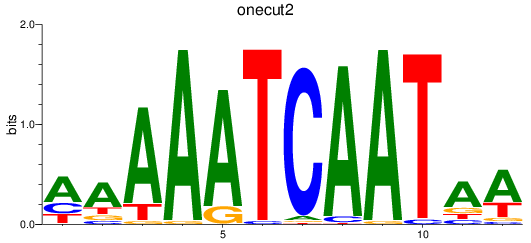
Results for onecut2
Z-value: 1.81
Transcription factors associated with onecut2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut2
|
ENSDARG00000090387 | one cut homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut2 | dr11_v1_chr21_-_1742159_1742159 | 0.79 | 1.1e-01 | Click! |
Activity profile of onecut2 motif
Sorted Z-values of onecut2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_3828560 | 1.96 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr1_-_21409877 | 1.93 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr25_-_9805269 | 1.48 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr25_+_7229046 | 1.38 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr5_+_27267186 | 1.37 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr18_+_50276653 | 1.35 |
ENSDART00000192120
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr4_+_26628822 | 1.32 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr4_-_5291256 | 1.31 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr18_+_50276337 | 1.23 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr8_-_23081511 | 1.21 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr3_-_28665291 | 1.18 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr17_-_45247332 | 1.13 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr18_-_37007061 | 1.11 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr9_+_44430705 | 1.09 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_+_20535869 | 1.08 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr18_-_37007294 | 1.06 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr9_+_44430974 | 1.04 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr4_-_8903240 | 1.04 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr3_-_28120092 | 1.03 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr24_+_25692802 | 1.02 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr18_+_642889 | 1.01 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr4_+_13810811 | 0.99 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr5_-_29643930 | 0.90 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr3_-_46811611 | 0.89 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr4_-_8902406 | 0.87 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr21_-_27881752 | 0.86 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr1_-_23157583 | 0.85 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr13_+_36144341 | 0.83 |
ENSDART00000182930
ENSDART00000187327 |
TTC9
|
si:ch211-259k16.3 |
| chr14_-_2348917 | 0.80 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr2_-_39759059 | 0.79 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr2_+_5563077 | 0.79 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr18_+_50890749 | 0.77 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr3_+_39853788 | 0.74 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr16_-_26255877 | 0.74 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr21_+_19070921 | 0.74 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr11_-_426525 | 0.74 |
ENSDART00000157054
|
znf831
|
zinc finger protein 831 |
| chr7_-_71434298 | 0.73 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr5_+_19309877 | 0.73 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr19_+_4916233 | 0.71 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr25_-_21085661 | 0.70 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr20_-_19422496 | 0.69 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr15_-_5467477 | 0.68 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr23_-_15216654 | 0.66 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr2_+_42871831 | 0.66 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr21_-_7687544 | 0.64 |
ENSDART00000134519
|
pde8b
|
phosphodiesterase 8B |
| chr2_-_31735142 | 0.62 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr22_-_7129631 | 0.62 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr15_-_26552652 | 0.60 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr1_+_16144615 | 0.59 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr25_+_19955598 | 0.58 |
ENSDART00000091547
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr8_+_35664152 | 0.58 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr9_+_44431174 | 0.58 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr19_-_41991104 | 0.56 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr5_+_61301525 | 0.56 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr8_+_9866351 | 0.55 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr19_-_17526735 | 0.54 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr18_-_25051846 | 0.54 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr17_-_14700889 | 0.54 |
ENSDART00000179975
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr5_-_65158203 | 0.53 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr4_-_4261673 | 0.53 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr15_+_19458982 | 0.52 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr8_-_2616326 | 0.47 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr17_+_43623598 | 0.47 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr11_+_43114108 | 0.46 |
ENSDART00000013642
ENSDART00000190266 |
foxg1b
|
forkhead box G1b |
| chr17_-_8169774 | 0.46 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr15_-_26552393 | 0.46 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr18_-_44847855 | 0.45 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr16_+_5612547 | 0.45 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr15_-_31265375 | 0.45 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr12_-_25217217 | 0.44 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr15_+_17446796 | 0.44 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr19_-_32641725 | 0.43 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr19_+_19512515 | 0.43 |
ENSDART00000180065
|
jazf1a
|
JAZF zinc finger 1a |
| chr2_-_32505091 | 0.42 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr17_-_14701529 | 0.42 |
ENSDART00000185142
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr23_+_25832689 | 0.41 |
ENSDART00000138907
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr10_-_15405564 | 0.40 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr23_-_3721444 | 0.38 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr4_-_2868112 | 0.38 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr11_-_22605981 | 0.37 |
ENSDART00000186923
|
myog
|
myogenin |
| chr18_+_36769758 | 0.37 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr21_+_22878834 | 0.37 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr18_-_21806613 | 0.37 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr21_+_22878991 | 0.35 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr2_-_17044959 | 0.35 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr5_+_4366431 | 0.34 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr4_-_9173552 | 0.33 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr16_-_13612650 | 0.32 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr6_-_16456093 | 0.31 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr7_-_49651378 | 0.31 |
ENSDART00000015040
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr21_-_43482426 | 0.31 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr5_+_7393346 | 0.30 |
ENSDART00000164238
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr19_-_18855513 | 0.30 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr19_-_41472228 | 0.30 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr5_-_14211487 | 0.30 |
ENSDART00000135731
|
npffr1l2
|
neuropeptide FF receptor 1 like 2 |
| chr19_-_18855717 | 0.30 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr18_+_36770166 | 0.29 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr2_+_45300512 | 0.29 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr23_+_28770225 | 0.29 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr14_+_7932973 | 0.28 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr3_-_8399774 | 0.27 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr21_+_3901775 | 0.27 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr14_-_41243180 | 0.27 |
ENSDART00000184548
|
drp2
|
dystrophin related protein 2 |
| chr23_-_19434199 | 0.27 |
ENSDART00000132543
|
klhdc8b
|
kelch domain containing 8B |
| chr17_+_21760032 | 0.25 |
ENSDART00000190425
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr18_+_14529005 | 0.25 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr20_-_6196989 | 0.24 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr18_-_20560007 | 0.24 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr3_-_60142530 | 0.23 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr4_+_7827261 | 0.23 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr6_-_8498676 | 0.23 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr22_-_38360205 | 0.22 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr15_-_35252522 | 0.22 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr6_-_10828880 | 0.22 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr7_+_25036188 | 0.22 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr6_-_8498908 | 0.22 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_-_9249943 | 0.21 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr15_+_21947117 | 0.21 |
ENSDART00000156234
|
ttc12
|
tetratricopeptide repeat domain 12 |
| chr17_-_25737452 | 0.21 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr25_+_18964782 | 0.20 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr23_+_12454542 | 0.20 |
ENSDART00000182259
ENSDART00000193043 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr5_+_34549365 | 0.19 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr7_-_41693004 | 0.18 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr2_-_6262441 | 0.17 |
ENSDART00000092190
|
arl14
|
ADP-ribosylation factor-like 14 |
| chr2_+_22042745 | 0.17 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr25_-_7753207 | 0.17 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr13_-_24877577 | 0.17 |
ENSDART00000182705
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr13_+_22316746 | 0.16 |
ENSDART00000188968
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr1_+_45958904 | 0.15 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr2_-_32387441 | 0.14 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr8_-_621142 | 0.14 |
ENSDART00000187576
ENSDART00000190316 |
si:ch211-87m7.2
|
si:ch211-87m7.2 |
| chr17_+_8175998 | 0.13 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr11_+_37201483 | 0.13 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr6_+_46431848 | 0.13 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr19_+_41006975 | 0.12 |
ENSDART00000138555
ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr20_+_54274431 | 0.12 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr9_+_7732714 | 0.11 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr21_+_20229926 | 0.10 |
ENSDART00000025860
|
si:dkey-247m21.3
|
si:dkey-247m21.3 |
| chr3_-_30434016 | 0.10 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr7_+_30933867 | 0.10 |
ENSDART00000156325
|
tjp1a
|
tight junction protein 1a |
| chr10_+_10801719 | 0.10 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr5_-_23675222 | 0.09 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr4_-_13255700 | 0.09 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr17_-_43623356 | 0.09 |
ENSDART00000075596
ENSDART00000136431 |
rtkn2a
|
rhotekin 2a |
| chr7_+_33372680 | 0.09 |
ENSDART00000193436
ENSDART00000099988 |
glceb
|
glucuronic acid epimerase b |
| chr12_+_26467847 | 0.08 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr11_+_5817202 | 0.08 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr5_-_28102698 | 0.08 |
ENSDART00000138140
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr15_-_30984557 | 0.08 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr17_+_6957042 | 0.07 |
ENSDART00000103839
|
zgc:172341
|
zgc:172341 |
| chr14_-_1313480 | 0.07 |
ENSDART00000097748
|
il21
|
interleukin 21 |
| chr7_+_42206847 | 0.06 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr6_-_49861506 | 0.06 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr17_-_14451405 | 0.06 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr8_+_40477264 | 0.06 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr8_-_37263524 | 0.06 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr20_-_6812688 | 0.05 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr8_-_14484599 | 0.05 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr19_-_27391179 | 0.05 |
ENSDART00000181108
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr4_-_76488854 | 0.05 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr21_+_4116437 | 0.04 |
ENSDART00000167791
ENSDART00000123759 |
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr20_+_22130284 | 0.04 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr24_-_37955993 | 0.04 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr8_-_45279411 | 0.04 |
ENSDART00000175207
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr23_+_24598910 | 0.04 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr10_-_24391716 | 0.04 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr13_-_40141630 | 0.04 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr5_+_34549845 | 0.03 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr16_-_16701718 | 0.03 |
ENSDART00000143550
|
si:dkey-8k3.2
|
si:dkey-8k3.2 |
| chr21_-_22812417 | 0.03 |
ENSDART00000079151
ENSDART00000112175 |
trpc6a
|
transient receptor potential cation channel, subfamily C, member 6a |
| chr23_-_20002459 | 0.03 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr2_-_42958619 | 0.03 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr19_+_42886413 | 0.02 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr20_-_40755614 | 0.01 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr7_-_68419830 | 0.01 |
ENSDART00000191606
|
CU468723.2
|
|
| chr5_+_37406358 | 0.00 |
ENSDART00000162811
|
klhl13
|
kelch-like family member 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 0.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.7 | GO:1900136 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 1.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.5 | GO:0060220 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.4 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 1.9 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 2.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 1.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.4 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.9 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.7 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 1.4 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.3 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.0 | 1.2 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 1.0 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.9 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.6 | GO:0044224 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.7 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.9 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 0.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 2.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 1.0 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 0.5 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 3.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.4 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 0.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |