Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
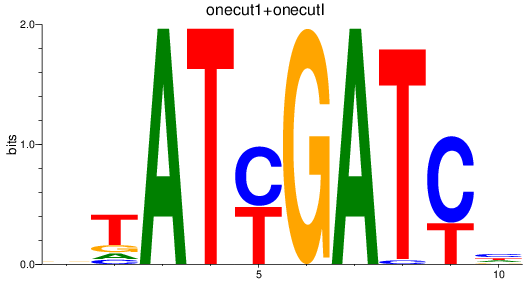
Results for onecut1+onecutl
Z-value: 2.85
Transcription factors associated with onecut1+onecutl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut1
|
ENSDARG00000007982 | one cut homeobox 1 |
|
onecutl
|
ENSDARG00000040253 | one cut domain, family member, like |
|
onecut1
|
ENSDARG00000112251 | one cut homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecutl | dr11_v1_chr16_-_29528198_29528198 | 0.89 | 4.2e-02 | Click! |
| onecut1 | dr11_v1_chr18_+_38755023_38755024 | 0.77 | 1.3e-01 | Click! |
Activity profile of onecut1+onecutl motif
Sorted Z-values of onecut1+onecutl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_7885707 | 3.03 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr8_-_31919624 | 2.48 |
ENSDART00000085573
|
rgs7bpa
|
regulator of G protein signaling 7 binding protein a |
| chr8_-_19051906 | 2.47 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr25_+_4837915 | 2.16 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr19_-_9829965 | 2.16 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr13_+_16521898 | 2.10 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr21_-_27881752 | 2.09 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr2_+_35240485 | 2.02 |
ENSDART00000179804
|
tnr
|
tenascin R (restrictin, janusin) |
| chr15_-_19677511 | 2.02 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr10_+_1638876 | 2.00 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr3_-_12890670 | 1.95 |
ENSDART00000159934
ENSDART00000188607 |
btbd17b
|
BTB (POZ) domain containing 17b |
| chr2_+_35240764 | 1.95 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr21_-_41305748 | 1.93 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr10_+_7182168 | 1.93 |
ENSDART00000172766
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr21_+_9628854 | 1.92 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr12_+_24344611 | 1.89 |
ENSDART00000093094
|
nrxn1a
|
neurexin 1a |
| chr8_+_7740004 | 1.89 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr16_-_44649053 | 1.88 |
ENSDART00000184807
|
CR925804.2
|
|
| chr7_+_23580082 | 1.78 |
ENSDART00000183245
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr20_+_3108597 | 1.75 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr10_+_25222367 | 1.71 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr15_-_25392589 | 1.71 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr23_+_28648864 | 1.70 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr9_-_18424844 | 1.68 |
ENSDART00000154351
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr24_-_17444067 | 1.62 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr6_+_55174744 | 1.60 |
ENSDART00000023562
|
SYT2
|
synaptotagmin 2 |
| chr25_-_11088839 | 1.60 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr17_-_20979077 | 1.59 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr5_+_55626693 | 1.58 |
ENSDART00000168908
ENSDART00000161412 |
ntrk2b
|
neurotrophic tyrosine kinase, receptor, type 2b |
| chr13_+_16522608 | 1.58 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr3_-_18711288 | 1.58 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr1_-_21409877 | 1.58 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr9_-_35557397 | 1.56 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr15_+_26399538 | 1.55 |
ENSDART00000159589
|
rtn4rl1b
|
reticulon 4 receptor-like 1b |
| chr12_+_24344963 | 1.54 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr7_-_22132265 | 1.52 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr6_+_3828560 | 1.52 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr10_+_37137464 | 1.52 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr11_+_25111846 | 1.50 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr15_-_24869826 | 1.50 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr4_+_10365857 | 1.49 |
ENSDART00000138890
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr9_+_22631672 | 1.48 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr24_+_24461558 | 1.42 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr12_-_3756405 | 1.42 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr12_-_4781801 | 1.40 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr19_-_867071 | 1.40 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr14_+_22172047 | 1.40 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr19_+_25649626 | 1.39 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr3_-_28665291 | 1.39 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_12767318 | 1.39 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr19_-_9882821 | 1.38 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr25_+_19954576 | 1.38 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr16_+_5184402 | 1.36 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr2_+_38608290 | 1.35 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr15_-_18429550 | 1.35 |
ENSDART00000136208
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr5_-_23429228 | 1.32 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr6_-_38419318 | 1.32 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr7_+_30875273 | 1.31 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr3_-_23407720 | 1.31 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_-_13886952 | 1.30 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr8_+_23916647 | 1.28 |
ENSDART00000143152
|
cpne5a
|
copine Va |
| chr17_-_16133249 | 1.28 |
ENSDART00000030919
|
pnoca
|
prepronociceptin a |
| chr7_+_73649686 | 1.27 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr16_+_21738194 | 1.24 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr19_+_37701450 | 1.22 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr17_+_8183393 | 1.17 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr13_+_35746440 | 1.15 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr12_+_39685485 | 1.13 |
ENSDART00000163403
|
LO017650.1
|
|
| chr3_-_23406964 | 1.13 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr5_-_29750377 | 1.12 |
ENSDART00000051474
|
barhl1a
|
BarH-like homeobox 1a |
| chr16_+_30301539 | 1.12 |
ENSDART00000186018
|
LO017848.1
|
|
| chr7_+_32722227 | 1.11 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr9_+_46644633 | 1.10 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr18_-_26510545 | 1.09 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr6_-_32349153 | 1.09 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr20_-_44575103 | 1.08 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr1_+_20069535 | 1.08 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr21_+_9576176 | 1.07 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr4_-_4507761 | 1.07 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr7_-_52842007 | 1.06 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr24_-_30091937 | 1.06 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr21_-_17482465 | 1.04 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr15_+_47418565 | 1.04 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr15_-_26686908 | 1.04 |
ENSDART00000185254
|
pitpnm3
|
PITPNM family member 3 |
| chr20_+_405811 | 1.03 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr9_+_32358514 | 1.03 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr19_-_8880688 | 1.02 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr24_-_21674950 | 1.01 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr15_-_5467477 | 1.01 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr23_-_44494401 | 1.01 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr16_-_24518027 | 1.00 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr5_+_3501859 | 0.98 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr3_+_43467595 | 0.98 |
ENSDART00000188300
|
galr2b
|
galanin receptor 2b |
| chr19_+_27479563 | 0.97 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr19_+_9533008 | 0.95 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr2_-_31735142 | 0.95 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr17_-_35076730 | 0.94 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr25_-_3979288 | 0.94 |
ENSDART00000157117
|
myrf
|
myelin regulatory factor |
| chr9_+_11034314 | 0.94 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr20_-_39273987 | 0.91 |
ENSDART00000127173
|
clu
|
clusterin |
| chr5_-_63515210 | 0.90 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr9_+_22632126 | 0.90 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr16_+_10346277 | 0.89 |
ENSDART00000081092
|
si:dkeyp-77h1.4
|
si:dkeyp-77h1.4 |
| chr18_-_42313798 | 0.88 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr18_-_20816103 | 0.86 |
ENSDART00000143233
|
cttnbp2
|
cortactin binding protein 2 |
| chr5_-_52277643 | 0.85 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr1_+_19930520 | 0.85 |
ENSDART00000158344
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr17_-_10249095 | 0.81 |
ENSDART00000159963
|
sstr1a
|
somatostatin receptor 1a |
| chr14_-_2933185 | 0.80 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr1_+_2712956 | 0.79 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr13_+_31070181 | 0.78 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr7_+_33314925 | 0.78 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr14_+_8127893 | 0.77 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr4_+_20318127 | 0.77 |
ENSDART00000028856
ENSDART00000132909 |
cacna1c
|
calcium channel, voltage-dependent, L type, alpha 1C subunit |
| chr16_-_26435431 | 0.76 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr22_+_17359346 | 0.76 |
ENSDART00000145434
|
gpr52
|
G protein-coupled receptor 52 |
| chr7_-_54217547 | 0.76 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr1_-_44704261 | 0.76 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr15_-_26636826 | 0.76 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr7_-_12789251 | 0.75 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr1_+_49814942 | 0.75 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr16_+_55168245 | 0.75 |
ENSDART00000171109
|
eomesb
|
eomesodermin homolog b |
| chr2_-_2096055 | 0.75 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr3_-_47876427 | 0.75 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr18_-_44611252 | 0.74 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr15_+_17441734 | 0.74 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr3_-_55531450 | 0.72 |
ENSDART00000155376
|
tex2
|
testis expressed 2 |
| chr19_-_17526735 | 0.71 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr16_+_52105227 | 0.71 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr15_-_1003553 | 0.71 |
ENSDART00000154195
|
si:dkey-77f5.6
|
si:dkey-77f5.6 |
| chr19_+_27479838 | 0.70 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr12_-_25065916 | 0.69 |
ENSDART00000159167
|
socs5a
|
suppressor of cytokine signaling 5a |
| chr3_+_39853788 | 0.69 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr22_+_110158 | 0.68 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr16_-_39900665 | 0.68 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr8_-_16259063 | 0.67 |
ENSDART00000057590
|
dmrta2
|
DMRT-like family A2 |
| chr21_-_31143903 | 0.67 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr13_+_42406883 | 0.66 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_+_57725708 | 0.65 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr20_-_50014936 | 0.65 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr11_+_6819050 | 0.63 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr20_-_7000225 | 0.63 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr11_+_43114108 | 0.62 |
ENSDART00000013642
ENSDART00000190266 |
foxg1b
|
forkhead box G1b |
| chr1_-_25438737 | 0.62 |
ENSDART00000134470
|
fhdc1
|
FH2 domain containing 1 |
| chr9_+_54417141 | 0.61 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr2_+_21486529 | 0.61 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr18_-_12612699 | 0.61 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr18_-_37007061 | 0.61 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr20_-_20931197 | 0.60 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr16_-_21181128 | 0.60 |
ENSDART00000133403
|
pde11al
|
phosphodiesterase 11a, like |
| chr2_+_38881165 | 0.59 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr3_-_61205711 | 0.58 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr6_-_18739519 | 0.57 |
ENSDART00000170998
|
tnrc6c2
|
trinucleotide repeat containing 6C2 |
| chr7_+_72460911 | 0.57 |
ENSDART00000160682
ENSDART00000168532 |
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr18_-_37007294 | 0.57 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr25_-_31118923 | 0.56 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr17_-_50233493 | 0.56 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr11_-_17713987 | 0.55 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr14_+_11762991 | 0.54 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr18_-_370286 | 0.54 |
ENSDART00000162633
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr21_+_1324416 | 0.53 |
ENSDART00000176557
|
TCF4
|
transcription factor 4 |
| chr16_-_40727455 | 0.53 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr9_+_36356740 | 0.52 |
ENSDART00000139033
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr15_-_14375452 | 0.52 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr16_+_23960744 | 0.52 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr2_+_35806672 | 0.52 |
ENSDART00000137384
|
rasal2
|
RAS protein activator like 2 |
| chr7_+_28006096 | 0.51 |
ENSDART00000173843
|
tub
|
tubby bipartite transcription factor |
| chr12_+_38770654 | 0.50 |
ENSDART00000155367
|
kif19
|
kinesin family member 19 |
| chr13_-_33000649 | 0.50 |
ENSDART00000133677
|
rbm25a
|
RNA binding motif protein 25a |
| chr9_+_37754845 | 0.50 |
ENSDART00000100592
|
pdia5
|
protein disulfide isomerase family A, member 5 |
| chr8_-_40555340 | 0.49 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr16_-_46393154 | 0.49 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr24_-_35707552 | 0.49 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr1_+_41498697 | 0.48 |
ENSDART00000114230
|
dtx4
|
deltex 4, E3 ubiquitin ligase |
| chr2_+_56463167 | 0.48 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr5_-_22952156 | 0.48 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr14_-_33083539 | 0.48 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr1_+_44710955 | 0.48 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr6_-_4214297 | 0.47 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr12_+_22407852 | 0.47 |
ENSDART00000178840
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr10_-_309894 | 0.46 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr16_+_25116827 | 0.45 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr20_-_39273505 | 0.45 |
ENSDART00000153114
|
clu
|
clusterin |
| chr1_-_10071422 | 0.45 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr5_+_19165949 | 0.44 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr10_+_44057502 | 0.44 |
ENSDART00000183868
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr2_+_45300512 | 0.43 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr6_+_37754763 | 0.43 |
ENSDART00000110770
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_18873113 | 0.42 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr8_-_22157301 | 0.42 |
ENSDART00000158383
|
nphp4
|
nephronophthisis 4 |
| chr16_+_23960933 | 0.42 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr5_+_50788329 | 0.42 |
ENSDART00000050979
|
fam81b
|
family with sequence similarity 81, member B |
| chr10_+_44057177 | 0.42 |
ENSDART00000164610
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr15_-_23442891 | 0.41 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr20_+_20731052 | 0.41 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr7_-_31938938 | 0.40 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr5_-_201600 | 0.40 |
ENSDART00000158495
|
CABZ01088906.1
|
|
| chr18_+_3498989 | 0.40 |
ENSDART00000172373
|
ompa
|
olfactory marker protein a |
| chr4_+_77920666 | 0.40 |
ENSDART00000129523
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr3_-_55649319 | 0.39 |
ENSDART00000176127
|
axin2
|
axin 2 (conductin, axil) |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut1+onecutl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.5 | 2.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 1.6 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 1.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.7 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.3 | 1.6 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.3 | 0.9 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 3.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 0.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 1.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.3 | 1.0 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 2.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 0.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 3.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 3.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.7 | GO:0046552 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.8 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.2 | 0.8 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 1.5 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 4.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.9 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 1.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.6 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 0.3 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.3 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.9 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 1.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.3 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.6 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 4.3 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.1 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 1.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 3.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 1.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.4 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.8 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.0 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.6 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.4 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.7 | GO:0006475 | internal protein amino acid acetylation(GO:0006475) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 2.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 6.1 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) negative regulation of systemic arterial blood pressure(GO:0003085) vasodilation(GO:0042311) |
| 0.0 | 0.7 | GO:0035336 | long-chain fatty acid metabolic process(GO:0001676) long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 1.6 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 1.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.5 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 3.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.8 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 6.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 2.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 8.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.5 | 1.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 3.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 1.4 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.3 | 1.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 3.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.7 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 3.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 0.6 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 2.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 1.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 2.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.3 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 4.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 2.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.7 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 2.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 1.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |