Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
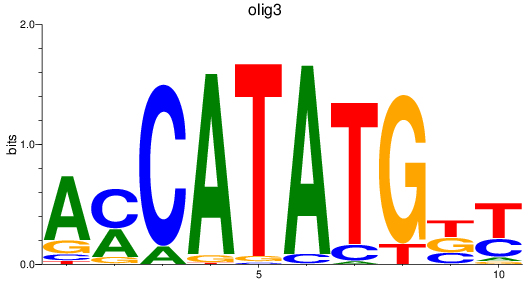
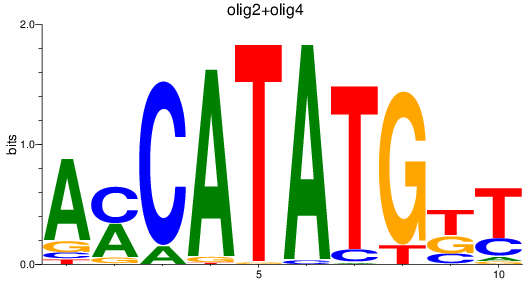
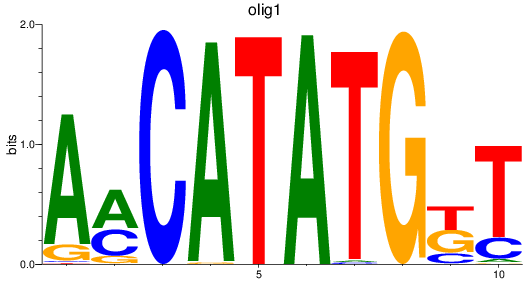
Results for olig3_olig2+olig4_olig1
Z-value: 0.67
Transcription factors associated with olig3_olig2+olig4_olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
olig3
|
ENSDARG00000074253 | oligodendrocyte transcription factor 3 |
|
olig2
|
ENSDARG00000040946 | oligodendrocyte lineage transcription factor 2 |
|
olig4
|
ENSDARG00000052610 | oligodendrocyte transcription factor 4 |
|
olig4
|
ENSDARG00000116678 | oligodendrocyte transcription factor 4 |
|
olig1
|
ENSDARG00000040948 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| olig4 | dr11_v1_chr13_+_45967179_45967179 | 0.49 | 4.1e-01 | Click! |
| olig1 | dr11_v1_chr9_-_32730487_32730487 | 0.45 | 4.4e-01 | Click! |
| olig2 | dr11_v1_chr9_-_32753535_32753535 | 0.36 | 5.5e-01 | Click! |
Activity profile of olig3_olig2+olig4_olig1 motif
Sorted Z-values of olig3_olig2+olig4_olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_1518360 | 0.41 |
ENSDART00000182788
|
c8b
|
complement component 8, beta polypeptide |
| chr22_-_24818066 | 0.33 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr21_+_6751405 | 0.30 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr8_-_39952727 | 0.27 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr15_+_36457888 | 0.26 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr18_+_13164325 | 0.26 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr14_-_25599002 | 0.26 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr16_+_50289916 | 0.25 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr22_-_18116635 | 0.25 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr2_-_17947389 | 0.24 |
ENSDART00000190089
ENSDART00000191872 ENSDART00000184039 ENSDART00000179791 |
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr15_-_21165237 | 0.23 |
ENSDART00000157069
|
A2ML1 (1 of many)
|
si:ch211-212c13.8 |
| chr2_-_39036604 | 0.22 |
ENSDART00000129963
|
rbp1
|
retinol binding protein 1b, cellular |
| chr4_-_8882572 | 0.22 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr23_+_26491931 | 0.21 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr3_+_40856095 | 0.21 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr10_-_23358357 | 0.21 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr2_-_27775236 | 0.21 |
ENSDART00000187983
|
XKR4
|
zgc:123035 |
| chr23_+_21544227 | 0.21 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr20_-_4031475 | 0.21 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr8_+_39998467 | 0.20 |
ENSDART00000073782
ENSDART00000134452 |
ggt5a
|
gamma-glutamyltransferase 5a |
| chr7_-_24022340 | 0.19 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr21_+_11415224 | 0.19 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr9_+_32859967 | 0.19 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr6_+_12865137 | 0.19 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr5_+_52625975 | 0.19 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr14_-_4682114 | 0.18 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr22_+_15979430 | 0.18 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr2_-_27774783 | 0.18 |
ENSDART00000161864
|
XKR4
|
zgc:123035 |
| chr22_-_36519590 | 0.18 |
ENSDART00000129318
|
CABZ01045212.1
|
|
| chr23_+_4299887 | 0.18 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr15_+_22267847 | 0.18 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr23_-_11870962 | 0.17 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr1_-_16507812 | 0.17 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr7_-_31938938 | 0.17 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr2_-_36040820 | 0.17 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr13_+_18202703 | 0.17 |
ENSDART00000109642
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr18_-_40708537 | 0.17 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr21_+_11468642 | 0.16 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr13_+_11439486 | 0.16 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr14_+_25817246 | 0.16 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr13_+_23677949 | 0.15 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr7_-_38633867 | 0.15 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr20_+_18994059 | 0.15 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr14_+_36521553 | 0.15 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr12_-_14922955 | 0.15 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr17_-_43594864 | 0.14 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr10_+_22012389 | 0.14 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr5_-_27994679 | 0.14 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr25_+_37397031 | 0.14 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr8_-_54223316 | 0.14 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr22_-_24757785 | 0.14 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr4_+_15968483 | 0.14 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr18_+_50278858 | 0.14 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr20_-_1265562 | 0.14 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr16_+_23972126 | 0.14 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr25_-_26753196 | 0.14 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr5_+_20148671 | 0.14 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr6_+_27151940 | 0.14 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr14_+_25816874 | 0.13 |
ENSDART00000005499
|
glra1
|
glycine receptor, alpha 1 |
| chr9_+_42095220 | 0.13 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr14_-_4556896 | 0.13 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr11_+_36158134 | 0.13 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr25_+_3994823 | 0.13 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr13_+_40019001 | 0.13 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr9_-_374693 | 0.13 |
ENSDART00000166571
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr23_-_21453614 | 0.13 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr22_-_16034137 | 0.13 |
ENSDART00000062629
|
zbtb37
|
zinc finger and BTB domain containing 37 |
| chr7_+_39634873 | 0.13 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr1_+_29759678 | 0.13 |
ENSDART00000054059
ENSDART00000101856 |
cpb2
|
carboxypeptidase B2 (plasma) |
| chr25_-_5740334 | 0.13 |
ENSDART00000169622
ENSDART00000168720 |
LO017739.1
|
|
| chr2_-_58640644 | 0.12 |
ENSDART00000192164
|
CABZ01081294.1
|
|
| chr7_-_24520866 | 0.12 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr9_+_38168012 | 0.12 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr12_+_24562667 | 0.12 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr24_-_30096666 | 0.12 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr19_+_11984725 | 0.12 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr20_+_54333774 | 0.12 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr17_-_15189397 | 0.12 |
ENSDART00000133710
ENSDART00000110507 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr7_-_71486162 | 0.12 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr23_+_20563779 | 0.12 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr21_+_31150438 | 0.11 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr11_+_5588122 | 0.11 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr16_+_23984755 | 0.11 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr8_+_50150834 | 0.11 |
ENSDART00000056074
|
entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr17_-_23727978 | 0.11 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr19_+_12801940 | 0.11 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr5_-_58173627 | 0.11 |
ENSDART00000159087
|
drd2b
|
dopamine receptor D2b |
| chr1_-_43712120 | 0.11 |
ENSDART00000074600
|
SLC9B2
|
si:dkey-162b23.4 |
| chr21_-_39546737 | 0.11 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr2_-_39017838 | 0.11 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr2_-_22966076 | 0.11 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr5_+_25952340 | 0.11 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr20_+_25486021 | 0.11 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr16_-_8132742 | 0.11 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr21_-_26918901 | 0.11 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr23_-_31648026 | 0.11 |
ENSDART00000133569
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_+_9277327 | 0.10 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr1_-_54706039 | 0.10 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr13_+_31285660 | 0.10 |
ENSDART00000148352
|
gdf10a
|
growth differentiation factor 10a |
| chr14_-_47314011 | 0.10 |
ENSDART00000178523
|
fstl5
|
follistatin-like 5 |
| chr1_-_29747702 | 0.10 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr21_+_31150773 | 0.10 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr15_+_15856178 | 0.10 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr12_-_17863467 | 0.10 |
ENSDART00000042006
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr13_+_771403 | 0.10 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr20_+_18994465 | 0.10 |
ENSDART00000152778
|
tdh
|
L-threonine dehydrogenase |
| chr25_-_11057753 | 0.10 |
ENSDART00000186551
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr5_-_2752573 | 0.10 |
ENSDART00000168121
|
lzts3a
|
leucine zipper, putative tumor suppressor family member 3a |
| chr18_-_25645680 | 0.10 |
ENSDART00000135874
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr5_+_50869091 | 0.10 |
ENSDART00000083294
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr5_-_35301800 | 0.10 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr18_+_38192499 | 0.10 |
ENSDART00000191849
|
nucb2b
|
nucleobindin 2b |
| chr17_-_42218652 | 0.10 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr20_+_34913069 | 0.10 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr15_+_4988189 | 0.10 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr16_+_5156420 | 0.10 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr9_+_32860345 | 0.10 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr2_+_24177006 | 0.09 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr10_-_32524771 | 0.09 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr5_-_25576462 | 0.09 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr8_-_37391372 | 0.09 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr4_-_4507761 | 0.09 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr22_+_4707663 | 0.09 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr13_+_36770738 | 0.09 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr10_-_6775271 | 0.09 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr23_+_7548797 | 0.09 |
ENSDART00000006765
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr22_+_38194151 | 0.09 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr14_-_2327825 | 0.09 |
ENSDART00000180328
ENSDART00000191135 ENSDART00000114302 ENSDART00000189869 |
pcdh2ab8
pcdh2aa1
|
protocadherin 2 alpha b 8 protocadherin 2 alpha a 1 |
| chr20_-_34028967 | 0.09 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr25_+_4787431 | 0.09 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr19_-_27578929 | 0.09 |
ENSDART00000177368
|
si:dkeyp-46h3.3
|
si:dkeyp-46h3.3 |
| chr14_-_46442459 | 0.09 |
ENSDART00000158770
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr12_+_31735159 | 0.09 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr4_+_33654247 | 0.09 |
ENSDART00000192537
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr3_-_47876427 | 0.09 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr5_-_43682930 | 0.09 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr25_-_8916913 | 0.09 |
ENSDART00000104629
ENSDART00000131748 |
furinb
|
furin (paired basic amino acid cleaving enzyme) b |
| chr6_+_48618512 | 0.09 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr2_+_24177190 | 0.09 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr1_-_29758947 | 0.09 |
ENSDART00000049514
ENSDART00000183571 ENSDART00000140345 |
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr4_+_4272622 | 0.08 |
ENSDART00000164810
|
ano2
|
anoctamin 2 |
| chr7_-_18598661 | 0.08 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr2_+_24770435 | 0.08 |
ENSDART00000078854
|
mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr17_-_722218 | 0.08 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr23_+_44665230 | 0.08 |
ENSDART00000144958
|
camta2
|
calmodulin binding transcription activator 2 |
| chr1_-_22803147 | 0.08 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr10_+_38813389 | 0.08 |
ENSDART00000136928
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr25_+_37130450 | 0.08 |
ENSDART00000183358
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr6_-_36182115 | 0.08 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr10_-_26226228 | 0.08 |
ENSDART00000182967
|
fhdc3
|
FH2 domain containing 3 |
| chr25_+_3294150 | 0.08 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr17_+_15534815 | 0.08 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr25_+_19954576 | 0.08 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr6_-_13308813 | 0.08 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr3_-_24909245 | 0.08 |
ENSDART00000178321
|
SHISA8
|
shisa family member 8 |
| chr14_+_31618982 | 0.08 |
ENSDART00000026195
|
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr24_+_37598441 | 0.08 |
ENSDART00000125145
|
rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr15_+_36115955 | 0.08 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr3_-_30625219 | 0.08 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr9_+_42066030 | 0.08 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr18_+_30878196 | 0.08 |
ENSDART00000099326
ENSDART00000146041 |
mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr7_-_71434298 | 0.08 |
ENSDART00000180507
|
lgi2a
|
leucine-rich repeat LGI family, member 2a |
| chr5_-_16983336 | 0.08 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr1_-_22652424 | 0.08 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr8_-_8489685 | 0.08 |
ENSDART00000131849
ENSDART00000064113 |
abt1
|
activator of basal transcription 1 |
| chr2_+_50608099 | 0.08 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr14_-_552036 | 0.07 |
ENSDART00000171317
|
SPATA5
|
spermatogenesis associated 5 |
| chr13_+_33368503 | 0.07 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr9_-_30363770 | 0.07 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr17_+_28882977 | 0.07 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr1_-_51684941 | 0.07 |
ENSDART00000113853
|
prokr1a
|
prokineticin receptor 1a |
| chr20_+_52442870 | 0.07 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr22_-_11054244 | 0.07 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr19_+_10661520 | 0.07 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr25_+_7423770 | 0.07 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr19_+_1184878 | 0.07 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr17_-_12389259 | 0.07 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr10_+_32560667 | 0.07 |
ENSDART00000182278
|
map6a
|
microtubule-associated protein 6a |
| chr5_+_24156170 | 0.07 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr21_+_13387965 | 0.07 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr20_+_66857 | 0.07 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr25_-_29273208 | 0.07 |
ENSDART00000148940
|
hcn4l
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4l |
| chr20_-_46467280 | 0.07 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr2_-_44282796 | 0.07 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr14_+_48964628 | 0.07 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr2_+_56937548 | 0.07 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr10_+_5689510 | 0.07 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_+_72630369 | 0.07 |
ENSDART00000170698
|
FO905040.1
|
|
| chr11_-_35975026 | 0.07 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr9_-_16109001 | 0.07 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr22_+_35275206 | 0.07 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr14_-_30960470 | 0.07 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr17_+_6956696 | 0.07 |
ENSDART00000171368
|
zgc:172341
|
zgc:172341 |
| chr19_+_43563179 | 0.07 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr19_+_38167468 | 0.07 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr1_-_51465730 | 0.07 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr9_+_22929675 | 0.07 |
ENSDART00000061299
|
tsn
|
translin |
| chr20_+_28803642 | 0.07 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr23_+_42304602 | 0.07 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr25_+_17339814 | 0.07 |
ENSDART00000141311
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of olig3_olig2+olig4_olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.2 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 0.3 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0050951 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.0 | GO:1900193 | regulation of oocyte maturation(GO:1900193) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.5 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0048914 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.0 | GO:0071514 | genetic imprinting(GO:0071514) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.0 | 0.0 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |