Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
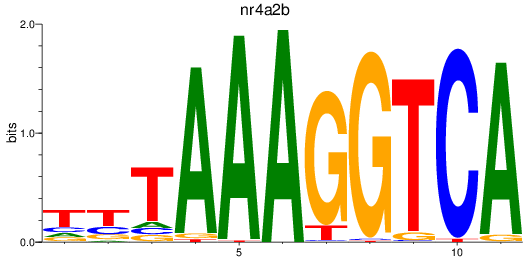
Results for nr4a2b
Z-value: 1.54
Transcription factors associated with nr4a2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a2b
|
ENSDARG00000044532 | nuclear receptor subfamily 4, group A, member 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a2b | dr11_v1_chr6_+_12462079_12462079 | 0.65 | 2.4e-01 | Click! |
Activity profile of nr4a2b motif
Sorted Z-values of nr4a2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_40150612 | 1.65 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr11_+_11201096 | 1.31 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr11_+_1409622 | 1.25 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr3_+_39566999 | 1.18 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_-_39764995 | 1.17 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_31871830 | 1.12 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr14_-_17121676 | 1.03 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr5_-_68093169 | 1.02 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr6_-_39765546 | 0.99 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr24_+_34089977 | 0.95 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr2_-_48753873 | 0.95 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr12_-_26415499 | 0.89 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr2_-_32457919 | 0.87 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr3_+_30257582 | 0.85 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr12_-_26423439 | 0.76 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr7_+_25913225 | 0.73 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr12_+_14084291 | 0.62 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr15_+_20403903 | 0.62 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr22_+_30184039 | 0.62 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr6_-_21189295 | 0.61 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr8_-_17997845 | 0.59 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr4_-_4932619 | 0.59 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr9_+_31795343 | 0.59 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr7_+_38588866 | 0.58 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr24_+_25471196 | 0.56 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr9_-_21067971 | 0.50 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr6_+_13206516 | 0.47 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr1_-_30689004 | 0.44 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr25_-_16969922 | 0.41 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr21_+_5080789 | 0.41 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr8_-_979735 | 0.39 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr3_+_31177972 | 0.38 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr6_-_10835849 | 0.37 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr7_+_34297271 | 0.35 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr16_-_25233515 | 0.35 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr10_+_41668483 | 0.34 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr5_+_36611128 | 0.34 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr7_+_22702437 | 0.33 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr3_-_15154871 | 0.33 |
ENSDART00000147704
|
si:dkey-282h22.5
|
si:dkey-282h22.5 |
| chr7_+_34296789 | 0.32 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_+_1278092 | 0.31 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr8_-_14052349 | 0.30 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr19_+_19652439 | 0.30 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr5_-_26330313 | 0.29 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr17_+_30545895 | 0.29 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr10_-_17745345 | 0.29 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr1_-_23110740 | 0.28 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr9_+_24065855 | 0.28 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr17_-_12389259 | 0.28 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr14_+_33882973 | 0.27 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr1_+_16127825 | 0.27 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr14_+_30340251 | 0.27 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr12_+_36891483 | 0.26 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr12_-_850457 | 0.25 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr23_+_24272421 | 0.25 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr13_-_21739142 | 0.24 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr19_-_703898 | 0.24 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr18_+_30511230 | 0.23 |
ENSDART00000078888
|
dusp22a
|
dual specificity phosphatase 22a |
| chr7_-_19146925 | 0.23 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr9_+_30633184 | 0.22 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr5_-_13076779 | 0.22 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr13_-_27660955 | 0.22 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr17_-_1705013 | 0.22 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr7_+_20524064 | 0.22 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr3_-_22212764 | 0.22 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr21_-_22724980 | 0.21 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr3_-_18189283 | 0.21 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr7_+_38667066 | 0.21 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr2_+_34767171 | 0.20 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr1_-_46875493 | 0.20 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr15_+_11840311 | 0.20 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr18_+_9382847 | 0.20 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr4_-_8060962 | 0.20 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr8_+_13106760 | 0.20 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr5_-_67721368 | 0.19 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr25_+_16214854 | 0.19 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr1_-_40102836 | 0.19 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr2_+_6885852 | 0.19 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr3_-_54500354 | 0.19 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr10_-_20524387 | 0.19 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr10_+_11265387 | 0.17 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr12_+_46708920 | 0.17 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr14_+_35023923 | 0.17 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr7_-_31794878 | 0.17 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr7_+_22702225 | 0.17 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr15_+_33691455 | 0.17 |
ENSDART00000170329
|
hdac12
|
histone deacetylase 12 |
| chr8_+_20825987 | 0.16 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr5_+_29652198 | 0.16 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr18_+_27926839 | 0.15 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr15_-_6946286 | 0.15 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr11_-_21363834 | 0.15 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr12_-_47648538 | 0.15 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr10_-_20524592 | 0.14 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr3_-_46818001 | 0.14 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr5_+_29652513 | 0.13 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr7_+_42206847 | 0.13 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr3_+_24190207 | 0.13 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr3_+_17806213 | 0.12 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr3_-_46817838 | 0.12 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_+_30368145 | 0.12 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr24_-_36802891 | 0.11 |
ENSDART00000088130
|
dcakd
|
dephospho-CoA kinase domain containing |
| chr5_+_24156170 | 0.11 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr5_-_16983336 | 0.11 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr3_-_28258462 | 0.11 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_-_24046999 | 0.11 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr20_-_10120442 | 0.11 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr14_+_30413758 | 0.10 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr6_-_29288155 | 0.09 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr24_+_39211288 | 0.09 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr7_-_31794476 | 0.09 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr6_-_41135215 | 0.09 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr2_-_41562868 | 0.09 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr6_+_29288223 | 0.09 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr1_+_44826367 | 0.09 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr23_-_45504991 | 0.09 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr24_-_24848612 | 0.09 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr4_-_2975461 | 0.09 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_44484853 | 0.09 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr18_+_13182528 | 0.09 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr8_-_44004135 | 0.08 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr17_+_30546579 | 0.08 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr6_-_50404732 | 0.08 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr1_-_58868306 | 0.08 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr21_-_43527198 | 0.07 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr6_-_35738836 | 0.07 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr18_+_31410652 | 0.07 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr9_+_35860975 | 0.06 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr10_+_7564106 | 0.06 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr5_-_33959868 | 0.06 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr19_+_32158010 | 0.06 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr21_-_41369370 | 0.06 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr8_+_53120278 | 0.05 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr6_+_29288006 | 0.05 |
ENSDART00000043496
|
zgc:172121
|
zgc:172121 |
| chr5_+_59494079 | 0.05 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr11_-_44999858 | 0.05 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr8_-_7440364 | 0.05 |
ENSDART00000180659
|
hdac6
|
histone deacetylase 6 |
| chr17_-_17764801 | 0.05 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr4_+_11723852 | 0.05 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr10_+_7563755 | 0.05 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr17_+_7595356 | 0.05 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr5_+_61738276 | 0.04 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr13_+_21768447 | 0.04 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr5_+_6670945 | 0.04 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr18_-_42333428 | 0.04 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr2_-_11912347 | 0.04 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr2_-_44971551 | 0.04 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr3_+_23687909 | 0.04 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr1_+_38153944 | 0.03 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr5_-_66301142 | 0.03 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr24_-_27256673 | 0.03 |
ENSDART00000181182
|
zmynd11
|
zinc finger, MYND-type containing 11 |
| chr24_-_1021318 | 0.03 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr8_-_22542467 | 0.03 |
ENSDART00000182588
ENSDART00000134542 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr8_-_42238543 | 0.02 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr24_+_26997798 | 0.02 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr23_-_28141419 | 0.02 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr22_-_11833317 | 0.02 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr22_+_31821815 | 0.02 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr5_-_57879138 | 0.02 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr12_-_29305220 | 0.01 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr1_+_20593653 | 0.01 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr24_+_22022109 | 0.01 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr18_+_27515640 | 0.01 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr18_-_2433011 | 0.00 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.0 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 2.2 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0043455 | regulation of secondary metabolic process(GO:0043455) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.3 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.1 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 2.2 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |