Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
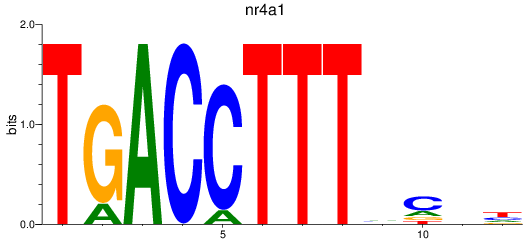
Results for nr4a1
Z-value: 1.46
Transcription factors associated with nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a1
|
ENSDARG00000000796 | nuclear receptor subfamily 4, group A, member 1 |
|
nr4a1
|
ENSDARG00000109473 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a1 | dr11_v1_chr23_-_32156278_32156278 | 0.23 | 7.0e-01 | Click! |
Activity profile of nr4a1 motif
Sorted Z-values of nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_1278092 | 1.64 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr10_-_17745345 | 1.60 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr6_-_31348999 | 1.50 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr8_-_14052349 | 1.27 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr4_-_7212875 | 1.06 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr4_+_12615836 | 1.04 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr2_+_34767171 | 1.01 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr14_+_14225048 | 0.87 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr25_+_3525494 | 0.86 |
ENSDART00000169775
|
RAB19 (1 of many)
|
RAB19, member RAS oncogene family |
| chr21_+_10076203 | 0.85 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr12_+_21525496 | 0.79 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr1_-_23110740 | 0.79 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr14_-_32258759 | 0.79 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr12_+_24562667 | 0.77 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr3_-_22212764 | 0.77 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr5_+_24156170 | 0.77 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr2_-_11912347 | 0.74 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr12_+_9817440 | 0.73 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr5_-_26330313 | 0.70 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr14_+_14224730 | 0.70 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr6_-_53204808 | 0.69 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr7_+_44484853 | 0.69 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr22_-_16997475 | 0.66 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr13_-_21739142 | 0.66 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr20_-_10120442 | 0.63 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr6_-_35738836 | 0.63 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr9_-_43142636 | 0.60 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr24_-_24848612 | 0.59 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr22_-_16997246 | 0.59 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr19_+_2275019 | 0.58 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr5_-_26323137 | 0.58 |
ENSDART00000133823
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr5_+_59449762 | 0.58 |
ENSDART00000150230
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr18_-_42333428 | 0.56 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr5_-_50600512 | 0.55 |
ENSDART00000190318
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr18_+_27515640 | 0.55 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr7_-_19146925 | 0.55 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr14_+_36246726 | 0.54 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr21_+_23953181 | 0.54 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr18_+_27926839 | 0.53 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr22_+_30184039 | 0.53 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr7_+_20524064 | 0.52 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr5_+_19448078 | 0.51 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr9_-_30346279 | 0.50 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr6_-_10320676 | 0.50 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr7_+_34620418 | 0.49 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr7_+_31132588 | 0.49 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr12_-_33568174 | 0.49 |
ENSDART00000142329
|
tdrkh
|
tudor and KH domain containing |
| chr5_+_62611400 | 0.48 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr11_-_6974022 | 0.48 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr5_+_24282570 | 0.47 |
ENSDART00000041905
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr6_-_45995401 | 0.45 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr24_+_4977862 | 0.44 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr7_+_34297271 | 0.44 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr14_+_36628131 | 0.43 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr10_+_7563755 | 0.43 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr3_-_37351225 | 0.40 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr8_+_51084993 | 0.40 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr5_+_65493699 | 0.40 |
ENSDART00000161567
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr20_-_27711970 | 0.40 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr3_+_33745014 | 0.39 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr7_+_34296789 | 0.39 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr23_-_24488696 | 0.39 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr22_-_15587360 | 0.38 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr8_+_39634114 | 0.38 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr7_-_42206720 | 0.38 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr10_+_7564106 | 0.38 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr6_-_41135215 | 0.38 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr11_-_17713987 | 0.37 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr8_-_45835056 | 0.36 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr22_-_11833317 | 0.36 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr10_+_41668483 | 0.34 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr19_+_3056450 | 0.34 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr3_+_39566999 | 0.34 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr5_+_10046643 | 0.33 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr2_-_8688759 | 0.32 |
ENSDART00000010257
|
miga1
|
mitoguardin 1 |
| chr21_+_7900107 | 0.31 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr1_-_50838160 | 0.31 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr6_-_39764995 | 0.31 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr18_+_9382847 | 0.31 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr17_-_47305034 | 0.30 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr8_+_10404310 | 0.30 |
ENSDART00000185573
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr11_+_1409622 | 0.30 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr3_-_41292275 | 0.30 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr6_+_29288223 | 0.30 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr24_+_4978055 | 0.30 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr8_-_15991801 | 0.29 |
ENSDART00000132005
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr8_-_45834825 | 0.29 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr20_+_40150612 | 0.28 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr12_-_48127985 | 0.28 |
ENSDART00000193805
|
npffr1
|
neuropeptide FF receptor 1 |
| chr21_-_43527198 | 0.28 |
ENSDART00000126092
|
irs4a
|
insulin receptor substrate 4a |
| chr7_+_15871408 | 0.27 |
ENSDART00000014572
|
pax6b
|
paired box 6b |
| chr13_+_22479988 | 0.27 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_30257582 | 0.27 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr18_-_2433011 | 0.26 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr3_+_32842825 | 0.26 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr2_-_42827336 | 0.25 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr15_-_11341635 | 0.25 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr20_-_14206698 | 0.25 |
ENSDART00000082441
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr8_-_15991473 | 0.24 |
ENSDART00000151746
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr20_-_26060154 | 0.24 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr6_+_13206516 | 0.23 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr2_-_4787566 | 0.23 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr10_+_24678092 | 0.22 |
ENSDART00000159270
|
thsd1
|
thrombospondin, type I, domain containing 1 |
| chr17_-_7371564 | 0.22 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr23_+_30730121 | 0.21 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr13_-_41155472 | 0.21 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr6_+_40952031 | 0.21 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr4_-_2975461 | 0.20 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_+_515181 | 0.20 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr5_+_41322783 | 0.20 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr24_-_8214253 | 0.20 |
ENSDART00000160432
|
CABZ01077956.1
|
|
| chr21_-_30648106 | 0.20 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr7_+_56253914 | 0.19 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr7_+_65398161 | 0.18 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr7_+_32021669 | 0.18 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr11_+_11201096 | 0.17 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr7_+_42206847 | 0.17 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr3_+_24190207 | 0.17 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr16_-_50203058 | 0.17 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr19_+_5146460 | 0.16 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr14_+_41576849 | 0.16 |
ENSDART00000074424
|
nitr13
|
novel immune-type receptor 13 |
| chr8_+_26083808 | 0.16 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr20_-_32981053 | 0.16 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr6_+_41957280 | 0.16 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr23_+_26003187 | 0.16 |
ENSDART00000147271
|
wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_+_30545895 | 0.15 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr15_+_1705167 | 0.15 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr10_+_36441124 | 0.15 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr3_-_45250924 | 0.15 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr3_-_51109286 | 0.14 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr6_-_39765546 | 0.14 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr17_+_450956 | 0.13 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr23_-_45504991 | 0.13 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr11_-_44999858 | 0.13 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr13_+_29193649 | 0.13 |
ENSDART00000045951
|
unc5b
|
unc-5 netrin receptor B |
| chr24_+_22022109 | 0.13 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr6_+_29288006 | 0.12 |
ENSDART00000043496
|
zgc:172121
|
zgc:172121 |
| chr15_-_16946124 | 0.12 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr22_+_29113796 | 0.12 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr20_+_21391181 | 0.12 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr21_+_5080789 | 0.11 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr8_-_979735 | 0.11 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr1_+_31638274 | 0.11 |
ENSDART00000057885
|
fgf8b
|
fibroblast growth factor 8 b |
| chr17_+_41463942 | 0.10 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr23_-_9859989 | 0.09 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr9_-_43082945 | 0.09 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr7_-_31794878 | 0.08 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr7_-_24005268 | 0.08 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr15_+_40281693 | 0.07 |
ENSDART00000186565
|
gal3st2
|
galactose-3-O-sulfotransferase 2 |
| chr18_+_31410652 | 0.07 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr12_+_46708920 | 0.07 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr3_-_27915270 | 0.07 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr16_-_45288599 | 0.07 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr24_-_6628359 | 0.06 |
ENSDART00000169731
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr20_+_13883353 | 0.06 |
ENSDART00000188006
|
nek2
|
NIMA-related kinase 2 |
| chr2_-_41562868 | 0.06 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr5_+_23201370 | 0.06 |
ENSDART00000138123
|
tpcn1
|
two pore segment channel 1 |
| chr22_-_17595310 | 0.06 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr7_+_38588866 | 0.06 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr14_+_48045193 | 0.06 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr8_+_21114338 | 0.06 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr8_+_53120278 | 0.06 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr3_-_5829501 | 0.05 |
ENSDART00000091017
|
pkn1b
|
protein kinase N1b |
| chr8_+_21406769 | 0.05 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr20_+_46897504 | 0.04 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr13_+_22480496 | 0.04 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr25_+_29160102 | 0.04 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr14_-_48103207 | 0.04 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr25_+_6266009 | 0.03 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr2_+_43755274 | 0.03 |
ENSDART00000138620
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr17_+_32374876 | 0.03 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr14_-_17121676 | 0.03 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr25_-_31898552 | 0.03 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr16_+_19677484 | 0.02 |
ENSDART00000150588
|
tmem196b
|
transmembrane protein 196b |
| chr14_+_33882973 | 0.02 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr17_+_5800286 | 0.01 |
ENSDART00000175504
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr6_-_29288155 | 0.01 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr8_+_26125218 | 0.01 |
ENSDART00000145095
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr19_+_19652439 | 0.01 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr6_-_24392426 | 0.00 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.3 | 0.8 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 0.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 0.7 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 2.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.7 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 1.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.3 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.2 | GO:0060137 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.6 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.5 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.7 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 2.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.7 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.7 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.7 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.5 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |