Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
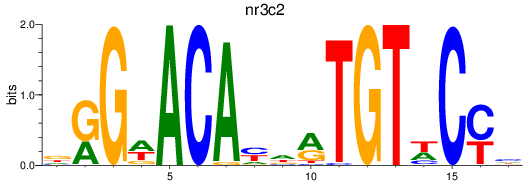
Results for nr3c2
Z-value: 0.62
Transcription factors associated with nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c2
|
ENSDARG00000102082 | nuclear receptor subfamily 3, group C, member 2 |
|
nr3c2
|
ENSDARG00000115513 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c2 | dr11_v1_chr1_-_37087966_37087966 | 0.13 | 8.3e-01 | Click! |
Activity profile of nr3c2 motif
Sorted Z-values of nr3c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41096058 | 0.85 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr8_+_7359294 | 0.58 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr6_+_41099787 | 0.58 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr5_-_43935119 | 0.46 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr16_+_50089417 | 0.46 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr5_-_43935460 | 0.45 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr24_-_38097305 | 0.36 |
ENSDART00000124321
|
crp2
|
C-reactive protein 2 |
| chr14_-_46113321 | 0.35 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr11_+_34522554 | 0.35 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr3_-_25275364 | 0.34 |
ENSDART00000163782
ENSDART00000145420 ENSDART00000133718 ENSDART00000055492 |
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr4_-_15420452 | 0.32 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr11_+_34523132 | 0.31 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr14_-_28001986 | 0.31 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr2_-_32512648 | 0.29 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr17_+_29345606 | 0.28 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr7_+_48288762 | 0.28 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr1_-_39909985 | 0.27 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr3_-_40051425 | 0.25 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr5_-_54792239 | 0.24 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_-_36519590 | 0.23 |
ENSDART00000129318
|
CABZ01045212.1
|
|
| chr15_-_18429550 | 0.23 |
ENSDART00000136208
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr6_-_6448519 | 0.20 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr15_+_40188076 | 0.19 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr7_+_58699718 | 0.18 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr8_-_50525360 | 0.18 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr23_+_44732863 | 0.16 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr11_-_34147205 | 0.16 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr11_-_43473824 | 0.16 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr15_+_37559570 | 0.16 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr5_-_31901468 | 0.16 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr3_-_37476475 | 0.15 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr24_-_20641000 | 0.11 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr16_+_11188810 | 0.10 |
ENSDART00000186011
|
cicb
|
capicua transcriptional repressor b |
| chr13_+_29510023 | 0.10 |
ENSDART00000187398
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr20_+_54336137 | 0.09 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr18_-_44611252 | 0.08 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_9417409 | 0.07 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr10_-_13178853 | 0.05 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr16_+_46430627 | 0.05 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr22_-_34979139 | 0.04 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr1_+_54199406 | 0.04 |
ENSDART00000176578
|
tsc2
|
TSC complex subunit 2 |
| chr8_-_45835056 | 0.03 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr4_+_11723852 | 0.02 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_-_45834825 | 0.02 |
ENSDART00000132965
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr25_-_3470910 | 0.02 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr1_-_28473350 | 0.02 |
ENSDART00000190608
ENSDART00000148175 |
si:ch1073-440b2.1
|
si:ch1073-440b2.1 |
| chr5_+_9382301 | 0.01 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr5_-_9540641 | 0.01 |
ENSDART00000124384
ENSDART00000160079 |
gak
|
cyclin G associated kinase |
| chr7_-_51749683 | 0.00 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |