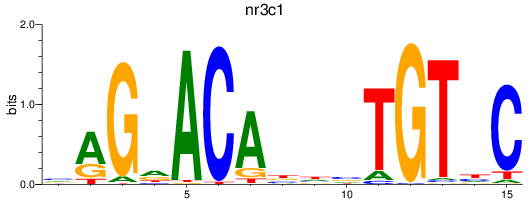
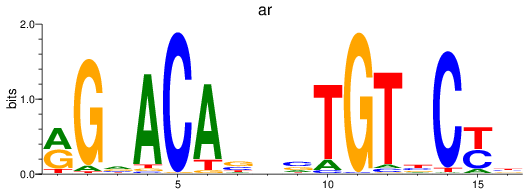
Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for nr3c1_ar
Z-value: 3.47
Transcription factors associated with nr3c1_ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c1
|
ENSDARG00000025032 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000112480 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
nr3c1
|
ENSDARG00000116957 | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
|
ar
|
ENSDARG00000067976 | androgen receptor |
|
ar
|
ENSDARG00000114287 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ar | dr11_v1_chr5_+_35561607_35561607 | -0.13 | 8.3e-01 | Click! |
| nr3c1 | dr11_v1_chr14_-_23799345_23799349 | -0.05 | 9.3e-01 | Click! |
Activity profile of nr3c1_ar motif
Sorted Z-values of nr3c1_ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41096058 | 5.75 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr6_+_41099787 | 4.94 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr21_+_20901505 | 2.74 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr8_+_7359294 | 2.58 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr12_+_13256415 | 2.53 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr16_+_50089417 | 2.52 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr7_+_48288762 | 2.40 |
ENSDART00000083569
|
oaz2b
|
ornithine decarboxylase antizyme 2b |
| chr1_-_26782573 | 2.37 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr16_+_10777116 | 2.20 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr17_-_6730247 | 2.19 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr3_-_50124413 | 1.99 |
ENSDART00000189920
|
cldnk
|
claudin k |
| chr7_+_6969909 | 1.99 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr21_-_30648106 | 1.95 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr15_+_40188076 | 1.93 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr13_-_226393 | 1.92 |
ENSDART00000172677
|
rtn4b
|
reticulon 4b |
| chr13_+_912123 | 1.91 |
ENSDART00000169931
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr16_+_10776688 | 1.90 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr20_+_28266892 | 1.87 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr12_-_46959990 | 1.65 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr2_-_155270 | 1.65 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr21_+_32338897 | 1.61 |
ENSDART00000110137
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr19_+_19412692 | 1.60 |
ENSDART00000113580
|
wu:fc38h03
|
wu:fc38h03 |
| chr7_-_32833153 | 1.60 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr4_+_4803698 | 1.60 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr5_-_43935119 | 1.57 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr1_-_59176949 | 1.54 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr23_-_1557195 | 1.51 |
ENSDART00000136436
|
epm2a
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_+_38556195 | 1.45 |
ENSDART00000138769
|
cdh24b
|
cadherin 24, type 2b |
| chr10_+_29431529 | 1.45 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr16_-_12266153 | 1.44 |
ENSDART00000113345
|
leng8
|
leukocyte receptor cluster (LRC) member 8 |
| chr5_-_43935460 | 1.41 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr18_+_17428258 | 1.41 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr10_-_24343507 | 1.39 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr25_+_31912153 | 1.39 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr15_-_9031996 | 1.36 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr20_+_34915945 | 1.35 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr4_-_789645 | 1.35 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr14_-_28001986 | 1.32 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr6_-_6254432 | 1.30 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr4_-_20511595 | 1.29 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr5_+_32206378 | 1.25 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr8_-_11202378 | 1.25 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr17_+_45737992 | 1.23 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr11_+_34522554 | 1.22 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr14_-_2355833 | 1.21 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr12_-_4683325 | 1.21 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr10_+_37500234 | 1.21 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr18_+_50278858 | 1.19 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr20_-_34801181 | 1.17 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr1_-_39909985 | 1.16 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr10_-_43655449 | 1.15 |
ENSDART00000099134
|
mef2ca
|
myocyte enhancer factor 2ca |
| chr3_-_25275364 | 1.14 |
ENSDART00000163782
ENSDART00000145420 ENSDART00000133718 ENSDART00000055492 |
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr18_-_35736591 | 1.13 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr3_-_58543658 | 1.13 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr15_+_37559570 | 1.11 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr17_+_29345606 | 1.11 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr11_+_34523132 | 1.10 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr3_-_5067585 | 1.09 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr14_-_46113321 | 1.09 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr4_-_11163112 | 1.08 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr15_-_18429550 | 1.08 |
ENSDART00000136208
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr19_-_32804535 | 1.08 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr6_-_6448519 | 1.05 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr5_+_19314574 | 1.03 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr15_-_5742531 | 1.02 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr10_+_34315719 | 1.02 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr19_+_8985230 | 1.01 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr21_+_25054420 | 0.97 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr16_+_32559821 | 0.97 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr23_-_31645760 | 0.97 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr11_+_37144328 | 0.95 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr11_+_6152643 | 0.94 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr24_-_24271629 | 0.90 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr4_+_19534833 | 0.89 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr4_-_15420452 | 0.89 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr15_-_23342752 | 0.86 |
ENSDART00000020425
|
mcamb
|
melanoma cell adhesion molecule b |
| chr21_+_30563115 | 0.86 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_-_131236 | 0.85 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr8_+_53452681 | 0.84 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr4_+_15968483 | 0.84 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr1_+_31674297 | 0.84 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr21_-_32462856 | 0.83 |
ENSDART00000147318
|
zgc:123105
|
zgc:123105 |
| chr22_+_12595144 | 0.82 |
ENSDART00000140054
ENSDART00000060979 ENSDART00000139826 |
zgc:92335
|
zgc:92335 |
| chr9_+_22677503 | 0.82 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr8_-_436137 | 0.81 |
ENSDART00000187021
|
CABZ01063543.1
|
|
| chr15_-_23647078 | 0.81 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr5_+_32162684 | 0.81 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr5_-_31901468 | 0.80 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr6_-_40006809 | 0.80 |
ENSDART00000085666
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr5_+_51909740 | 0.79 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr8_-_50525360 | 0.77 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr20_-_9273669 | 0.76 |
ENSDART00000175069
|
syt14b
|
synaptotagmin XIVb |
| chr21_+_32339158 | 0.75 |
ENSDART00000161723
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr6_+_40354424 | 0.74 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr10_-_15672862 | 0.73 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr18_-_19491162 | 0.73 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr10_-_13178853 | 0.72 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr1_+_24076243 | 0.72 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr4_-_11737939 | 0.70 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr20_-_29475172 | 0.69 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr19_-_32487469 | 0.68 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr5_+_65492183 | 0.68 |
ENSDART00000162804
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr5_-_23362602 | 0.67 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr8_-_39984593 | 0.67 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr6_-_40029423 | 0.66 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr16_-_22225295 | 0.66 |
ENSDART00000163519
|
LO017682.1
|
|
| chr2_+_25929619 | 0.65 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr18_+_17428506 | 0.65 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr25_-_12803723 | 0.65 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr23_+_44732863 | 0.65 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr3_-_40051425 | 0.64 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr12_+_49135755 | 0.64 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr14_+_46028003 | 0.64 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr14_-_2036604 | 0.63 |
ENSDART00000192446
|
BX005294.2
|
|
| chr23_+_23183449 | 0.63 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr7_+_18364176 | 0.62 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr10_+_41765944 | 0.62 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr9_-_43538328 | 0.62 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr17_+_25187670 | 0.62 |
ENSDART00000190873
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr4_-_2014406 | 0.62 |
ENSDART00000180463
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr3_-_35800221 | 0.61 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr25_-_3469576 | 0.60 |
ENSDART00000186738
|
hbp1
|
HMG-box transcription factor 1 |
| chr16_+_46430627 | 0.60 |
ENSDART00000127681
|
rpz6
|
rapunzel 6 |
| chr25_+_7423770 | 0.60 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr10_+_36439293 | 0.59 |
ENSDART00000043802
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr24_-_20641000 | 0.57 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr5_+_37649206 | 0.56 |
ENSDART00000149151
ENSDART00000097723 |
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr17_-_45247151 | 0.55 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr1_+_54199406 | 0.55 |
ENSDART00000176578
|
tsc2
|
TSC complex subunit 2 |
| chr6_-_17849786 | 0.55 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr10_-_43611643 | 0.55 |
ENSDART00000134953
ENSDART00000134219 ENSDART00000138962 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr19_+_30633453 | 0.55 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr14_+_22114918 | 0.55 |
ENSDART00000166610
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr2_-_32512648 | 0.54 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr5_-_41494831 | 0.53 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr3_-_36440705 | 0.53 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr21_+_11503212 | 0.53 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr1_-_1631399 | 0.52 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr23_+_9088191 | 0.52 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr25_-_29363934 | 0.51 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr21_+_1378250 | 0.51 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr24_+_32525146 | 0.50 |
ENSDART00000132417
ENSDART00000110185 |
yme1l1a
|
YME1-like 1a |
| chr5_-_54792239 | 0.50 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_3453589 | 0.50 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr18_+_16744307 | 0.49 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr18_-_44611252 | 0.49 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_65536095 | 0.49 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr19_-_41213718 | 0.49 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr15_-_13254480 | 0.48 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr20_-_16498991 | 0.48 |
ENSDART00000104137
|
ches1
|
checkpoint suppressor 1 |
| chr8_-_45835056 | 0.48 |
ENSDART00000022242
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr3_-_4663602 | 0.48 |
ENSDART00000083532
|
slc25a38a
|
solute carrier family 25, member 38a |
| chr21_-_25565392 | 0.47 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr16_+_47207691 | 0.47 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr2_+_3533117 | 0.45 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr19_+_12801940 | 0.45 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr18_+_45114392 | 0.45 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr24_+_18714212 | 0.45 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr7_-_35036770 | 0.45 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr4_-_2162688 | 0.44 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr6_+_60112200 | 0.44 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr21_-_32467099 | 0.44 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr2_-_12243213 | 0.43 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr6_-_22064158 | 0.43 |
ENSDART00000153585
|
cfap100
|
cilia and flagella associated protein 100 |
| chr25_-_6420800 | 0.43 |
ENSDART00000153768
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr6_+_49412754 | 0.43 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr10_-_45379831 | 0.43 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr7_+_21752168 | 0.42 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr12_-_18568834 | 0.42 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr23_-_33944597 | 0.42 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr12_+_32073660 | 0.42 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr8_+_54055390 | 0.42 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr23_+_35708730 | 0.42 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr16_+_18535618 | 0.42 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr1_+_7517454 | 0.41 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr14_-_32486757 | 0.41 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr10_+_21758811 | 0.41 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr18_+_30511230 | 0.41 |
ENSDART00000078888
|
dusp22a
|
dual specificity phosphatase 22a |
| chr16_+_25116827 | 0.41 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr23_+_33777519 | 0.41 |
ENSDART00000159642
|
CR381531.2
|
|
| chr23_+_6752828 | 0.41 |
ENSDART00000105179
|
zgc:158254
|
zgc:158254 |
| chr8_-_46700278 | 0.41 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr23_-_35064785 | 0.41 |
ENSDART00000172240
|
BX294434.1
|
|
| chr8_+_28467893 | 0.40 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr4_+_25651720 | 0.40 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr19_-_10730488 | 0.40 |
ENSDART00000126033
|
slc9a3.1
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 1 |
| chr17_-_277046 | 0.38 |
ENSDART00000182587
|
LO018437.1
|
|
| chr3_+_49397115 | 0.38 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr14_-_9982603 | 0.37 |
ENSDART00000054687
|
il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr23_+_20803270 | 0.37 |
ENSDART00000097381
|
zgc:154075
|
zgc:154075 |
| chr3_-_56896702 | 0.37 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr8_+_35172594 | 0.37 |
ENSDART00000177146
|
BX897670.1
|
|
| chr14_-_2270973 | 0.37 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr3_-_36602069 | 0.37 |
ENSDART00000165414
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr16_-_6424816 | 0.37 |
ENSDART00000164864
ENSDART00000141860 |
mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr2_-_3403020 | 0.37 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr5_+_51594209 | 0.36 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr25_-_5189989 | 0.36 |
ENSDART00000097520
|
shisal1b
|
shisa like 1b |
| chr21_+_28478663 | 0.36 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr16_-_22781446 | 0.36 |
ENSDART00000144107
|
lenep
|
lens epithelial protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c1_ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.5 | 1.9 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.4 | 2.4 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 10.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 0.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.6 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 0.9 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.3 | 4.8 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.7 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 1.6 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 2.2 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.2 | 1.6 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.5 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 1.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 1.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.4 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 1.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.6 | GO:1903405 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 1.8 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 2.4 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.1 | 2.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 3.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.7 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.6 | GO:0098900 | intestinal epithelial cell differentiation(GO:0060575) regulation of action potential(GO:0098900) |
| 0.1 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.4 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0051481 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.2 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 1.7 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.8 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.2 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0071230 | TORC1 signaling(GO:0038202) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 2.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 3.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.0 | GO:0035676 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.8 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.6 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.0 | 2.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.2 | 3.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 1.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 2.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 2.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.9 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 1.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 2.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 1.9 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 1.7 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.4 | 4.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.2 | 0.7 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.2 | 1.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 2.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.6 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.7 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.3 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.8 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 2.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |