Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
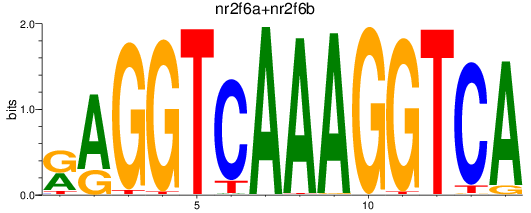
Results for nr2f6a+nr2f6b
Z-value: 0.27
Transcription factors associated with nr2f6a+nr2f6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f6b
|
ENSDARG00000003165 | nuclear receptor subfamily 2, group F, member 6b |
|
nr2f6a
|
ENSDARG00000003607 | nuclear receptor subfamily 2, group F, member 6a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f6a | dr11_v1_chr2_+_24374669_24374669 | 0.87 | 5.4e-02 | Click! |
| nr2f6b | dr11_v1_chr11_+_6116096_6116096 | -0.40 | 5.1e-01 | Click! |
Activity profile of nr2f6a+nr2f6b motif
Sorted Z-values of nr2f6a+nr2f6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_45128375 | 0.28 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr22_-_11833317 | 0.22 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr24_-_21172122 | 0.21 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr25_-_19420949 | 0.18 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr10_+_20128267 | 0.18 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr1_+_58353661 | 0.17 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr24_+_39137001 | 0.17 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr1_+_25848231 | 0.16 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr19_+_2275019 | 0.16 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr23_+_30730121 | 0.16 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr23_-_45504991 | 0.16 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr18_-_12957451 | 0.15 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr24_-_1021318 | 0.14 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr18_+_16330025 | 0.14 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr18_+_31410652 | 0.13 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr18_+_27515640 | 0.13 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr15_+_47386939 | 0.13 |
ENSDART00000128224
|
FO904873.1
|
|
| chr21_+_1119046 | 0.13 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr22_-_3595439 | 0.12 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr18_-_1185772 | 0.12 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr9_-_25255490 | 0.10 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr8_+_36803415 | 0.10 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr5_-_26330313 | 0.09 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr9_-_37613792 | 0.09 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr7_+_25000060 | 0.09 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr25_+_15647993 | 0.08 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr5_+_49744713 | 0.08 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr2_-_56649883 | 0.08 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr13_-_37127970 | 0.08 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr24_+_39034090 | 0.07 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr6_+_40952031 | 0.07 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr18_-_18850302 | 0.07 |
ENSDART00000131965
ENSDART00000167624 |
tgm2l
|
transglutaminase 2, like |
| chr12_+_46708920 | 0.06 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr25_+_20119466 | 0.06 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr12_+_26670778 | 0.06 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr2_-_41571454 | 0.06 |
ENSDART00000022643
|
znf622
|
zinc finger protein 622 |
| chr8_+_8012570 | 0.06 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr11_-_44999858 | 0.06 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr4_-_75172216 | 0.06 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr23_-_42810664 | 0.05 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr11_+_30729745 | 0.05 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr25_+_18587338 | 0.05 |
ENSDART00000180287
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr12_+_49125510 | 0.04 |
ENSDART00000185804
|
FO704607.1
|
|
| chr17_+_23926796 | 0.04 |
ENSDART00000021177
|
pex13
|
peroxisomal biogenesis factor 13 |
| chr2_-_22659450 | 0.04 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr24_-_7587401 | 0.04 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr23_+_36340520 | 0.04 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr23_-_46126444 | 0.04 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr22_-_16997887 | 0.04 |
ENSDART00000138233
|
nfia
|
nuclear factor I/A |
| chr19_-_3056235 | 0.04 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr23_+_2914577 | 0.04 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr7_-_36113098 | 0.03 |
ENSDART00000064913
|
fto
|
fat mass and obesity associated |
| chr25_+_22017182 | 0.03 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr7_-_24520866 | 0.03 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_-_7399767 | 0.03 |
ENSDART00000181689
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr22_-_16997475 | 0.03 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr3_+_1179601 | 0.03 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr9_+_32859967 | 0.03 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr9_+_34380299 | 0.03 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr1_+_58182400 | 0.03 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr23_-_21797517 | 0.02 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr4_-_858434 | 0.02 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr1_+_58303892 | 0.02 |
ENSDART00000147678
|
CR769768.1
|
|
| chr8_-_4097722 | 0.02 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr1_+_23783349 | 0.02 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr13_+_45980163 | 0.02 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr17_+_450956 | 0.02 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr12_+_48681601 | 0.02 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr7_+_48297842 | 0.02 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr6_+_40775800 | 0.01 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr15_+_47618221 | 0.01 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr22_-_16997246 | 0.01 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr17_-_25303486 | 0.01 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr23_+_27912079 | 0.01 |
ENSDART00000171859
|
BX539336.1
|
|
| chr12_-_31457301 | 0.01 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr1_+_58221372 | 0.01 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr23_+_44634187 | 0.01 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr1_+_58442694 | 0.01 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr21_-_44512893 | 0.01 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr17_-_6954719 | 0.01 |
ENSDART00000188180
|
zbtb24
|
zinc finger and BTB domain containing 24 |
| chr1_-_51720633 | 0.00 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr1_+_58260886 | 0.00 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr9_-_28990649 | 0.00 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr7_+_38267136 | 0.00 |
ENSDART00000173613
|
gpatch1
|
G patch domain containing 1 |
| chr3_-_41292275 | 0.00 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr4_-_16451375 | 0.00 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr8_+_8699085 | 0.00 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr9_-_15208129 | 0.00 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f6a+nr2f6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.0 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |