Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
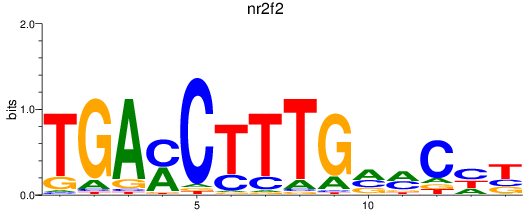
Results for nr2f2
Z-value: 2.20
Transcription factors associated with nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f2
|
ENSDARG00000040926 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f2 | dr11_v1_chr18_-_23875370_23875370 | -0.83 | 8.4e-02 | Click! |
Activity profile of nr2f2 motif
Sorted Z-values of nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_37022601 | 2.74 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr15_-_108414 | 2.51 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr10_-_322769 | 2.21 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr7_+_25036188 | 2.16 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr10_+_19569052 | 2.13 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr18_-_43866526 | 2.07 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr19_-_27564458 | 1.98 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr9_-_9982696 | 1.86 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr9_+_38292947 | 1.84 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr18_+_13162728 | 1.83 |
ENSDART00000101472
|
tat
|
tyrosine aminotransferase |
| chr19_+_9277327 | 1.71 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr22_-_24818066 | 1.68 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr17_-_2573021 | 1.68 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr10_-_21542702 | 1.68 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr17_-_2578026 | 1.65 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr3_-_3496738 | 1.61 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr12_-_6033824 | 1.60 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr17_-_2595736 | 1.60 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_-_31675669 | 1.51 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr23_-_44786844 | 1.48 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr22_-_23668356 | 1.47 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr18_-_43866001 | 1.45 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr22_+_15624371 | 1.45 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr3_-_36750068 | 1.44 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr17_-_30702411 | 1.42 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr17_-_2590222 | 1.41 |
ENSDART00000185711
|
CR759892.1
|
|
| chr5_-_3839285 | 1.37 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr3_+_12755535 | 1.34 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr20_-_43741159 | 1.30 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr20_+_2281933 | 1.28 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr3_+_12744083 | 1.26 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr19_-_3303995 | 1.26 |
ENSDART00000105150
|
si:ch211-133n4.9
|
si:ch211-133n4.9 |
| chr12_+_6041575 | 1.26 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr7_-_52096498 | 1.25 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr8_+_30709685 | 1.25 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr17_-_2584423 | 1.22 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr8_+_39802506 | 1.20 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr16_-_22294265 | 1.20 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr7_-_51953807 | 1.18 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr21_-_27443995 | 1.17 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr10_+_9561066 | 1.15 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr20_-_52967878 | 1.12 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr1_+_24387659 | 1.12 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr6_+_60055168 | 1.12 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr16_-_21785261 | 1.10 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr2_-_20120904 | 1.09 |
ENSDART00000186002
ENSDART00000124724 |
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr23_-_24488696 | 1.07 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr5_+_31605322 | 1.01 |
ENSDART00000135037
|
si:dkey-220k22.3
|
si:dkey-220k22.3 |
| chr19_-_3056235 | 1.00 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr3_+_12732382 | 1.00 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr16_+_23913943 | 0.99 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr18_+_40462445 | 0.96 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr25_+_18587338 | 0.94 |
ENSDART00000180287
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr22_-_7050 | 0.91 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr22_-_38274188 | 0.91 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr17_-_49407091 | 0.90 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr19_-_27564980 | 0.90 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr18_+_17611627 | 0.87 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr15_+_28268135 | 0.86 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr23_+_2740741 | 0.84 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr13_-_9886579 | 0.81 |
ENSDART00000101926
|
si:ch211-117n7.7
|
si:ch211-117n7.7 |
| chr6_+_60036767 | 0.81 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr17_+_48314724 | 0.80 |
ENSDART00000125617
|
smoc1
|
SPARC related modular calcium binding 1 |
| chr24_-_2843107 | 0.80 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr7_+_59649399 | 0.80 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr3_+_31680592 | 0.78 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr19_+_17386393 | 0.76 |
ENSDART00000034837
|
rpl15
|
ribosomal protein L15 |
| chr5_-_62317496 | 0.76 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr4_-_19693978 | 0.76 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr16_-_33001153 | 0.75 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr9_+_33216945 | 0.74 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr3_+_12718100 | 0.74 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr25_-_12803723 | 0.74 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr17_+_450956 | 0.72 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr20_-_36809059 | 0.71 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr20_-_25522911 | 0.71 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr16_+_11558868 | 0.69 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr22_+_2143107 | 0.69 |
ENSDART00000172339
ENSDART00000161280 ENSDART00000163671 ENSDART00000172090 ENSDART00000163573 |
znf1164
|
zinc finger protein 1164 |
| chr8_+_999421 | 0.68 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr3_+_12848123 | 0.68 |
ENSDART00000166689
|
si:ch211-8c17.4
|
si:ch211-8c17.4 |
| chr9_+_8380728 | 0.67 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr17_-_25737452 | 0.66 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr22_+_15331214 | 0.66 |
ENSDART00000136566
|
sult3st4
|
sulfotransferase family 3, cytosolic sulfotransferase 4 |
| chr14_+_31751260 | 0.66 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr25_+_418932 | 0.66 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr18_-_6534357 | 0.66 |
ENSDART00000192886
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr17_-_6613458 | 0.65 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr6_-_28117995 | 0.65 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr2_+_42715995 | 0.65 |
ENSDART00000143419
ENSDART00000183914 ENSDART00000184371 ENSDART00000185485 ENSDART00000037332 |
ftr12
|
finTRIM family, member 12 |
| chr21_-_1640547 | 0.63 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr3_+_34121156 | 0.63 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr24_-_30275204 | 0.63 |
ENSDART00000164187
|
snx7
|
sorting nexin 7 |
| chr11_+_6116096 | 0.63 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr25_-_35143360 | 0.62 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr19_-_9648542 | 0.62 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr7_-_24520866 | 0.61 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_+_5172848 | 0.61 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr6_+_27624023 | 0.61 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr3_-_31618037 | 0.61 |
ENSDART00000183547
|
moto
|
minamoto |
| chr9_+_21151138 | 0.60 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr10_-_22797959 | 0.59 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr25_+_34246625 | 0.59 |
ENSDART00000082578
|
bnip2
|
BCL2 interacting protein 2 |
| chr22_-_17595310 | 0.59 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr5_-_24124118 | 0.58 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr3_-_18805225 | 0.58 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr5_+_21211135 | 0.57 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr16_-_19568388 | 0.57 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr8_-_8698607 | 0.57 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr3_+_12710350 | 0.57 |
ENSDART00000157959
|
cyp2k18
|
cytochrome P450, family 2, subfamily K, polypeptide 18 |
| chr25_+_36348401 | 0.56 |
ENSDART00000103006
|
hist1h2a3
|
histone cluster 1 H2A family member 3 |
| chr19_+_48024457 | 0.55 |
ENSDART00000163823
|
kpnb1
|
karyopherin (importin) beta 1 |
| chr5_-_8171625 | 0.55 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr2_-_57076687 | 0.55 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr16_+_33143503 | 0.55 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr22_-_28668442 | 0.53 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr19_-_5332784 | 0.53 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr12_-_34580988 | 0.53 |
ENSDART00000152926
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr21_-_43992027 | 0.53 |
ENSDART00000188612
|
cdx1b
|
caudal type homeobox 1 b |
| chr12_+_2648043 | 0.52 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr2_-_55861351 | 0.52 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr4_-_16451375 | 0.52 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr3_+_58167288 | 0.52 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr4_-_36139585 | 0.51 |
ENSDART00000132071
|
znf992
|
zinc finger protein 992 |
| chr3_-_10677890 | 0.51 |
ENSDART00000155382
ENSDART00000171319 |
si:ch1073-144j5.2
|
si:ch1073-144j5.2 |
| chr7_-_71389375 | 0.51 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr6_+_27315173 | 0.51 |
ENSDART00000108831
|
espnla
|
espin like a |
| chr5_-_24238733 | 0.51 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr10_+_35526528 | 0.50 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr2_-_24407933 | 0.50 |
ENSDART00000088584
|
si:dkey-208k22.6
|
si:dkey-208k22.6 |
| chr4_+_74396786 | 0.50 |
ENSDART00000127501
ENSDART00000174347 |
zmp:0000001020
|
zmp:0000001020 |
| chr13_+_13668991 | 0.50 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr20_-_15090862 | 0.50 |
ENSDART00000063892
ENSDART00000122592 |
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr2_-_24289641 | 0.49 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr14_-_32876280 | 0.49 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr1_+_58094551 | 0.48 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr9_-_33121535 | 0.48 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr7_-_38183331 | 0.48 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr12_+_22870372 | 0.48 |
ENSDART00000182264
ENSDART00000191789 ENSDART00000130594 ENSDART00000190433 |
afap1
|
actin filament associated protein 1 |
| chr5_-_13564961 | 0.47 |
ENSDART00000146827
|
si:ch211-230g14.3
|
si:ch211-230g14.3 |
| chr11_-_30636163 | 0.47 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr21_+_33454147 | 0.47 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr19_-_47555956 | 0.47 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr4_-_31064105 | 0.47 |
ENSDART00000157670
|
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr12_-_46176115 | 0.46 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr25_+_34247107 | 0.46 |
ENSDART00000148507
|
bnip2
|
BCL2 interacting protein 2 |
| chr3_+_31662126 | 0.46 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr25_+_8316953 | 0.46 |
ENSDART00000154598
|
muc2.2
|
mucin 2.2, oligomeric mucus/gel-forming |
| chr17_+_21817002 | 0.45 |
ENSDART00000136727
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr13_+_8696825 | 0.45 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr5_+_19479200 | 0.45 |
ENSDART00000137703
|
aldh3b2
|
aldehyde dehydrogenase 3 family, member B2 |
| chr1_+_57050899 | 0.45 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr15_-_34418525 | 0.45 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr23_-_33738945 | 0.45 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr17_+_20173882 | 0.45 |
ENSDART00000155379
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr13_-_3516473 | 0.45 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr16_+_38360002 | 0.44 |
ENSDART00000087346
ENSDART00000148101 |
zgc:113232
|
zgc:113232 |
| chr17_-_12408109 | 0.44 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr9_+_54984537 | 0.44 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr5_-_36549024 | 0.44 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr11_+_6115621 | 0.44 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr8_-_8349653 | 0.43 |
ENSDART00000025214
|
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr9_-_46382637 | 0.43 |
ENSDART00000085738
|
lct
|
lactase |
| chr3_-_34561624 | 0.43 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr9_+_30464641 | 0.43 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr23_+_44633858 | 0.42 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr9_-_41153896 | 0.42 |
ENSDART00000059667
|
wdr75
|
WD repeat domain 75 |
| chr23_+_17437554 | 0.42 |
ENSDART00000184282
|
BX649300.3
|
|
| chr22_+_15308526 | 0.41 |
ENSDART00000188280
|
CU104797.1
|
|
| chr4_+_12031958 | 0.41 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr8_-_20243389 | 0.41 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr7_-_51775688 | 0.41 |
ENSDART00000149793
|
bmp15
|
bone morphogenetic protein 15 |
| chr2_-_42314218 | 0.41 |
ENSDART00000141210
ENSDART00000190495 |
ftr07
|
finTRIM family, member 7 |
| chr24_+_24808955 | 0.40 |
ENSDART00000080963
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr12_-_5120175 | 0.40 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr6_-_49873020 | 0.40 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr5_+_43870389 | 0.40 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr16_+_54875530 | 0.40 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr10_-_11385155 | 0.40 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr20_-_49650293 | 0.40 |
ENSDART00000151239
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr10_-_35410518 | 0.40 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr17_-_50374005 | 0.39 |
ENSDART00000149773
|
otofb
|
otoferlin b |
| chr2_+_3533458 | 0.39 |
ENSDART00000133007
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr3_-_4455951 | 0.39 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr5_-_7897316 | 0.38 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr22_-_910926 | 0.38 |
ENSDART00000180075
|
FP016205.1
|
|
| chr19_+_7604527 | 0.38 |
ENSDART00000132107
|
s100s
|
S100 calcium binding protein S |
| chr20_-_9658405 | 0.38 |
ENSDART00000023809
|
nid2b
|
nidogen 2b (osteonidogen) |
| chr7_-_72295181 | 0.38 |
ENSDART00000180598
|
muc5.3
|
mucin 5.3 |
| chr2_-_23569833 | 0.38 |
ENSDART00000138889
|
si:dkey-58b18.10
|
si:dkey-58b18.10 |
| chr19_-_25005609 | 0.38 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr1_+_56886214 | 0.37 |
ENSDART00000152718
ENSDART00000182408 |
si:ch211-1f22.8
|
si:ch211-1f22.8 |
| chr9_+_32860345 | 0.37 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr1_+_58470287 | 0.37 |
ENSDART00000160245
ENSDART00000169945 |
si:ch73-236c18.7
|
si:ch73-236c18.7 |
| chr1_-_53468160 | 0.37 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr21_-_22724980 | 0.36 |
ENSDART00000035469
|
c1qa
|
complement component 1, q subcomponent, A chain |
| chr9_+_32859967 | 0.36 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr5_-_10007897 | 0.36 |
ENSDART00000109052
|
CR936408.1
|
Danio rerio uncharacterized LOC799523 (LOC799523), mRNA. |
| chr16_-_1503023 | 0.36 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr12_-_5120339 | 0.36 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr11_+_30314885 | 0.36 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.6 | 3.4 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) |
| 0.5 | 2.3 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.4 | 6.1 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.8 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.3 | 1.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.3 | 2.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 0.7 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.7 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 1.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 0.8 | GO:0006660 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.1 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 7.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.7 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.4 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 0.4 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 1.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.8 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.9 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.6 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.2 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.7 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.1 | 0.3 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 0.2 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 1.7 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 1.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.4 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.8 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.0 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.9 | GO:0031047 | gene silencing by RNA(GO:0031047) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 4.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.8 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 6.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.6 | 6.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.5 | 2.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.5 | 1.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 2.7 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.3 | 1.2 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.3 | 1.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 1.7 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.2 | 1.1 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 0.7 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.5 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 8.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 3.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 1.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 3.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.0 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.9 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |