Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
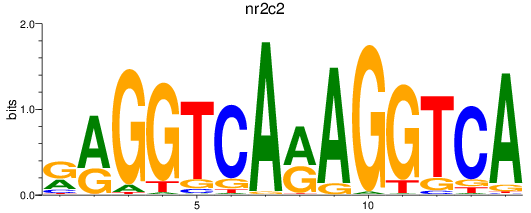
Results for nr2c2
Z-value: 0.91
Transcription factors associated with nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c2
|
ENSDARG00000042477 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c2 | dr11_v1_chr8_+_53120278_53120278 | 0.17 | 7.9e-01 | Click! |
Activity profile of nr2c2 motif
Sorted Z-values of nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_60036767 | 1.25 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr23_-_45705525 | 1.19 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr22_-_15587360 | 1.02 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr3_+_39566999 | 0.94 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_+_31871830 | 0.92 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr7_+_31838320 | 0.91 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr17_+_53424415 | 0.82 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr21_-_19006631 | 0.77 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr11_+_11201096 | 0.71 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr10_+_35417099 | 0.66 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr5_-_36837846 | 0.61 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr15_-_29598679 | 0.57 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr9_-_43082945 | 0.56 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr16_-_22713152 | 0.56 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr23_+_42810055 | 0.54 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr10_-_22797959 | 0.51 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr3_-_12187245 | 0.47 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr6_-_40722200 | 0.46 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr18_+_1615 | 0.44 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr11_+_77526 | 0.43 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr17_+_53425037 | 0.43 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr1_-_59240975 | 0.42 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr6_-_40722480 | 0.41 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr3_-_34561624 | 0.38 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr14_+_989733 | 0.38 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr6_-_14004772 | 0.37 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr6_+_1787160 | 0.37 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr1_+_58353661 | 0.37 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr3_+_27770110 | 0.37 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr17_+_15788100 | 0.36 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr17_+_33418475 | 0.35 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr9_+_35077546 | 0.35 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr8_+_15251448 | 0.35 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr16_-_24612871 | 0.35 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr6_+_27339962 | 0.35 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr23_+_19790962 | 0.34 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr3_-_18410968 | 0.34 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr7_+_24114694 | 0.34 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr25_+_15647993 | 0.33 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr6_-_49873020 | 0.32 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr7_+_24115082 | 0.32 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr11_-_6974022 | 0.31 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr3_+_301479 | 0.30 |
ENSDART00000165169
|
CABZ01075611.1
|
|
| chr23_-_44903048 | 0.30 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr8_+_31872992 | 0.30 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr22_-_31020690 | 0.30 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr3_-_32590164 | 0.30 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr5_+_22579975 | 0.30 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr19_+_2631565 | 0.29 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr21_-_308852 | 0.29 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr13_-_37127970 | 0.29 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr5_+_24282570 | 0.29 |
ENSDART00000041905
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr22_+_29994093 | 0.28 |
ENSDART00000104778
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr18_+_8917766 | 0.28 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr20_-_49117438 | 0.27 |
ENSDART00000057700
|
naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr16_-_25233515 | 0.27 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr20_+_15015557 | 0.27 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr5_+_9246018 | 0.26 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr1_+_58182400 | 0.26 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr14_-_29859067 | 0.26 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr1_-_47431453 | 0.26 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr10_-_29272707 | 0.26 |
ENSDART00000022605
ENSDART00000184594 ENSDART00000185371 |
tmem126a
|
transmembrane protein 126A |
| chr1_-_17797802 | 0.26 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr20_+_47953047 | 0.25 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr3_+_1179601 | 0.25 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr4_-_16833518 | 0.25 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr25_+_25766033 | 0.25 |
ENSDART00000103638
ENSDART00000039952 |
idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_+_58470287 | 0.24 |
ENSDART00000160245
ENSDART00000169945 |
si:ch73-236c18.7
|
si:ch73-236c18.7 |
| chr23_-_28808480 | 0.24 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr22_+_19486711 | 0.23 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr16_+_54875530 | 0.23 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr1_+_58221372 | 0.23 |
ENSDART00000138467
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr12_-_47782623 | 0.23 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr24_+_81527 | 0.23 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr4_-_991043 | 0.23 |
ENSDART00000184706
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr25_+_14694876 | 0.23 |
ENSDART00000050478
ENSDART00000188093 |
mpped2
|
metallophosphoesterase domain containing 2b |
| chr5_-_26330313 | 0.22 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr6_-_25165693 | 0.22 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr16_-_24605969 | 0.22 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr8_-_25329967 | 0.21 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr20_-_47953524 | 0.21 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr1_+_23783349 | 0.21 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr1_-_59232267 | 0.21 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr7_-_2090594 | 0.21 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr2_+_35880600 | 0.21 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr10_+_20128267 | 0.21 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr18_-_43866526 | 0.21 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr14_+_34501245 | 0.21 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr1_-_51720633 | 0.20 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr1_-_26664840 | 0.20 |
ENSDART00000102500
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr12_+_22870372 | 0.20 |
ENSDART00000182264
ENSDART00000191789 ENSDART00000130594 ENSDART00000190433 |
afap1
|
actin filament associated protein 1 |
| chr21_-_22317920 | 0.19 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr20_-_26937453 | 0.19 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr2_-_25143373 | 0.19 |
ENSDART00000160108
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr7_+_59649399 | 0.19 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr1_+_58312680 | 0.18 |
ENSDART00000138007
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr8_+_4745340 | 0.18 |
ENSDART00000182894
|
ufd1l
|
ubiquitin recognition factor in ER associated degradation 1 |
| chr7_+_24520518 | 0.18 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr8_-_979735 | 0.18 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr13_-_36566260 | 0.17 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr5_+_43870389 | 0.17 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr3_-_7134113 | 0.17 |
ENSDART00000180849
|
BX005085.6
|
|
| chr15_-_6946286 | 0.17 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr2_-_7666021 | 0.17 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr23_-_29394505 | 0.17 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr20_-_32270866 | 0.17 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr14_+_15331486 | 0.17 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr11_+_6116503 | 0.17 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr11_+_11751006 | 0.17 |
ENSDART00000180372
ENSDART00000081018 |
jupa
|
junction plakoglobin a |
| chr8_+_25629941 | 0.17 |
ENSDART00000186925
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr3_+_40289418 | 0.17 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr1_+_58119117 | 0.17 |
ENSDART00000146788
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr19_+_40861853 | 0.17 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr1_+_15062 | 0.16 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr14_+_30413758 | 0.16 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr23_+_17437554 | 0.16 |
ENSDART00000184282
|
BX649300.3
|
|
| chr6_+_39930825 | 0.16 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr3_-_19495814 | 0.16 |
ENSDART00000162248
|
CU571315.3
|
|
| chr14_-_48103207 | 0.16 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr23_-_10696626 | 0.16 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr24_-_2843107 | 0.16 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr1_+_57145072 | 0.16 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr25_-_35106751 | 0.16 |
ENSDART00000180682
ENSDART00000073457 |
zgc:194989
|
zgc:194989 |
| chr1_+_58139102 | 0.15 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr13_+_41819817 | 0.15 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr7_+_19374683 | 0.15 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr5_-_68927728 | 0.15 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr2_-_56649883 | 0.15 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr12_-_34580988 | 0.15 |
ENSDART00000152926
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr13_+_713941 | 0.15 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr9_-_32684008 | 0.15 |
ENSDART00000041751
|
ercc1
|
excision repair cross-complementation group 1 |
| chr4_-_75172216 | 0.15 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr16_-_19568388 | 0.14 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr20_+_27093042 | 0.14 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr16_-_5115993 | 0.14 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr10_+_589501 | 0.14 |
ENSDART00000188415
|
LO018557.1
|
|
| chr20_+_36806398 | 0.14 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr24_+_9590188 | 0.14 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr3_+_22432069 | 0.14 |
ENSDART00000123054
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr21_+_15704556 | 0.14 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr4_+_47656992 | 0.14 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr16_+_7985886 | 0.14 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr10_+_11265387 | 0.14 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr5_+_35786141 | 0.13 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr15_+_34963316 | 0.13 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr7_+_59649746 | 0.13 |
ENSDART00000181067
|
rpl34
|
ribosomal protein L34 |
| chr7_+_38267136 | 0.13 |
ENSDART00000173613
|
gpatch1
|
G patch domain containing 1 |
| chr21_+_31253048 | 0.13 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr3_+_1749793 | 0.13 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr16_+_28602423 | 0.13 |
ENSDART00000182350
|
crot
|
carnitine O-octanoyltransferase |
| chr20_-_34164278 | 0.13 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr17_-_8976307 | 0.13 |
ENSDART00000092113
|
zranb1b
|
zinc finger, RAN-binding domain containing 1b |
| chr1_-_54063520 | 0.13 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr1_+_58303892 | 0.13 |
ENSDART00000147678
|
CR769768.1
|
|
| chr14_+_30413312 | 0.13 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr14_-_29858883 | 0.13 |
ENSDART00000141034
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr10_+_24692076 | 0.13 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr5_-_30984010 | 0.13 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr1_+_58290933 | 0.13 |
ENSDART00000140536
|
si:dkey-222h21.6
|
si:dkey-222h21.6 |
| chr5_+_38276582 | 0.12 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_-_58868306 | 0.12 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr8_+_40275830 | 0.12 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr16_-_32128345 | 0.12 |
ENSDART00000102100
ENSDART00000137699 |
cnot3a
|
CCR4-NOT transcription complex, subunit 3a |
| chr11_-_29816429 | 0.12 |
ENSDART00000069132
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr6_-_40429411 | 0.12 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr7_-_24046999 | 0.12 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr4_+_34119824 | 0.12 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr20_-_51656512 | 0.12 |
ENSDART00000129965
|
LO018154.1
|
|
| chr7_-_19613388 | 0.12 |
ENSDART00000173664
|
si:ch211-212k18.13
|
si:ch211-212k18.13 |
| chr7_-_40578733 | 0.12 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr16_+_42830152 | 0.12 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_+_47618221 | 0.12 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr1_+_58260886 | 0.12 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr7_-_35393900 | 0.12 |
ENSDART00000173769
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr4_+_38981587 | 0.12 |
ENSDART00000142713
|
si:dkey-66k12.3
|
si:dkey-66k12.3 |
| chr1_+_58312187 | 0.11 |
ENSDART00000142285
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr21_-_1640547 | 0.11 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr4_-_50926767 | 0.11 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr7_+_34297271 | 0.11 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr19_-_9648542 | 0.11 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr14_-_34700633 | 0.11 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_+_34296789 | 0.11 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr8_+_2530065 | 0.11 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr14_+_97017 | 0.11 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr15_-_1844048 | 0.11 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_+_17865554 | 0.11 |
ENSDART00000181009
ENSDART00000162822 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr1_+_58150000 | 0.11 |
ENSDART00000137836
|
si:ch211-15j1.3
|
si:ch211-15j1.3 |
| chr6_+_42475730 | 0.11 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr18_-_127558 | 0.11 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr11_-_44999858 | 0.11 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr8_-_2529878 | 0.10 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr24_-_11446156 | 0.10 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr19_-_32600823 | 0.10 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr8_+_8699085 | 0.10 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr1_-_44314 | 0.10 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr13_+_45980163 | 0.10 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr20_+_46513651 | 0.10 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr5_-_32890807 | 0.10 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr17_-_22137199 | 0.10 |
ENSDART00000089689
|
mrps5
|
mitochondrial ribosomal protein S5 |
| chr7_-_8156169 | 0.10 |
ENSDART00000182960
ENSDART00000190184 ENSDART00000190885 ENSDART00000189867 ENSDART00000160836 |
si:cabz01030277.1
si:ch211-163c2.3
|
si:cabz01030277.1 si:ch211-163c2.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.2 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 0.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.2 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.1 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:1905133 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) heterochromatin assembly(GO:0031507) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.2 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.2 | 1.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.2 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.8 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.2 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |