Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
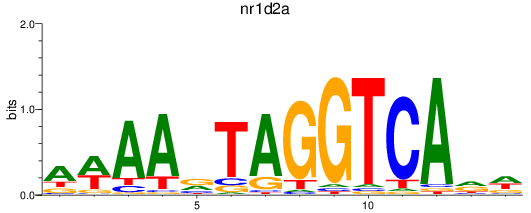
Results for nr1d2a
Z-value: 1.38
Transcription factors associated with nr1d2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1d2a
|
ENSDARG00000003820 | nuclear receptor subfamily 1, group D, member 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr1d2a | dr11_v1_chr16_+_50089417_50089417 | 0.71 | 1.8e-01 | Click! |
Activity profile of nr1d2a motif
Sorted Z-values of nr1d2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_70155935 | 2.02 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr17_-_6738538 | 1.92 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr7_+_40228422 | 1.53 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr18_-_39473055 | 1.52 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr6_+_57541776 | 1.46 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr23_+_19564392 | 1.40 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr3_+_35298078 | 1.37 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr17_-_15528597 | 1.29 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr25_+_6306885 | 1.26 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr3_-_2592350 | 1.24 |
ENSDART00000192325
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr6_-_39605734 | 1.23 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr3_-_2591942 | 1.19 |
ENSDART00000127971
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr14_-_42997145 | 1.18 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr18_+_22302635 | 1.13 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr2_-_16380283 | 1.10 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr12_+_28574863 | 1.09 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr20_+_16750177 | 1.06 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr8_-_33114202 | 1.05 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr5_-_11573490 | 1.05 |
ENSDART00000109577
|
FO704871.1
|
|
| chr5_-_67878064 | 1.03 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr2_+_20430366 | 0.99 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr22_-_38480186 | 0.99 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr22_-_11124419 | 0.97 |
ENSDART00000149634
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr1_-_44704261 | 0.95 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr18_-_42313798 | 0.95 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr9_-_48937089 | 0.91 |
ENSDART00000193442
|
cers6
|
ceramide synthase 6 |
| chr1_+_9708801 | 0.90 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr22_+_17828267 | 0.89 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr25_-_1235457 | 0.88 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr6_-_49159207 | 0.87 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr10_+_2232023 | 0.87 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr1_-_30039331 | 0.86 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr25_+_14165447 | 0.85 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_+_44692272 | 0.84 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr9_+_17982737 | 0.84 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr16_-_8927425 | 0.79 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr20_+_36233873 | 0.79 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr12_-_3756405 | 0.77 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr16_+_12632428 | 0.74 |
ENSDART00000184600
ENSDART00000180537 |
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr19_-_15335787 | 0.74 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr9_-_43207768 | 0.69 |
ENSDART00000192523
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr11_+_30672282 | 0.68 |
ENSDART00000023981
ENSDART00000188846 |
ttbk1a
|
tau tubulin kinase 1a |
| chr24_-_32408404 | 0.67 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr22_-_834106 | 0.67 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr8_+_21146262 | 0.67 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr18_+_907266 | 0.66 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr18_+_27738349 | 0.65 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr12_-_979789 | 0.64 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr17_-_42213285 | 0.64 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr3_+_32698424 | 0.63 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr15_+_2559875 | 0.63 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr1_+_19930520 | 0.58 |
ENSDART00000158344
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr10_-_25628555 | 0.56 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr12_+_12112384 | 0.55 |
ENSDART00000152431
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr1_-_49498116 | 0.54 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr1_-_21297748 | 0.54 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr8_+_43016714 | 0.53 |
ENSDART00000142671
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr12_+_19305390 | 0.52 |
ENSDART00000183987
ENSDART00000066391 |
csnk1e
|
casein kinase 1, epsilon |
| chr19_-_8732037 | 0.50 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr5_+_43470544 | 0.49 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr20_-_14680897 | 0.49 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr5_-_30079434 | 0.49 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr18_-_35736591 | 0.48 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr1_-_31140096 | 0.48 |
ENSDART00000172243
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr17_-_7371564 | 0.47 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr16_-_24561354 | 0.46 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr3_-_26190804 | 0.45 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr2_-_42558549 | 0.44 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr7_+_15871408 | 0.44 |
ENSDART00000014572
|
pax6b
|
paired box 6b |
| chr6_+_32415132 | 0.43 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr10_-_7988396 | 0.42 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr4_-_22519516 | 0.42 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr7_+_66048102 | 0.39 |
ENSDART00000104523
|
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr15_+_17100412 | 0.38 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr5_-_23179319 | 0.38 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr6_-_45995401 | 0.37 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr11_-_21303946 | 0.34 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr21_-_2209012 | 0.34 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr15_+_42599501 | 0.33 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr20_+_46620374 | 0.33 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr17_+_5793248 | 0.33 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr21_-_40557281 | 0.30 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr9_-_43213229 | 0.30 |
ENSDART00000139775
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr8_+_42917515 | 0.30 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr9_+_35860975 | 0.29 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr5_-_13778726 | 0.29 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr20_+_34320635 | 0.28 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr16_+_4838808 | 0.28 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr13_+_30172645 | 0.28 |
ENSDART00000137114
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr22_-_29689485 | 0.28 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr6_+_37752781 | 0.27 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr12_-_7234915 | 0.26 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr5_-_35888499 | 0.26 |
ENSDART00000193932
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr21_+_30549512 | 0.25 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr23_-_44233408 | 0.23 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr8_+_30452945 | 0.23 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr14_-_25111496 | 0.23 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr15_+_17100697 | 0.22 |
ENSDART00000183565
ENSDART00000123197 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr25_-_17918810 | 0.21 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr7_+_15871156 | 0.19 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr6_-_40768654 | 0.16 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr21_+_26606741 | 0.14 |
ENSDART00000002767
|
esrra
|
estrogen-related receptor alpha |
| chr12_-_32013125 | 0.14 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr23_+_21278948 | 0.14 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr8_+_21225064 | 0.14 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr17_+_49500820 | 0.13 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr17_-_23416897 | 0.13 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr23_+_44427188 | 0.12 |
ENSDART00000160207
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
| chr6_-_8244474 | 0.12 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr14_-_29859067 | 0.12 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr9_-_8661436 | 0.11 |
ENSDART00000130442
|
col4a1
|
collagen, type IV, alpha 1 |
| chr12_+_23912074 | 0.11 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr9_+_33267211 | 0.11 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr10_-_1788376 | 0.11 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_35733335 | 0.11 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr2_-_56655769 | 0.11 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr22_+_786556 | 0.10 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr9_-_43213057 | 0.09 |
ENSDART00000059448
ENSDART00000133589 |
sestd1
|
SEC14 and spectrin domains 1 |
| chr17_-_29271359 | 0.08 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr2_-_38337122 | 0.08 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr21_-_32781612 | 0.08 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr7_+_71586485 | 0.08 |
ENSDART00000165582
|
smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr19_+_24896409 | 0.08 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr22_-_15704704 | 0.07 |
ENSDART00000017838
ENSDART00000130238 |
safb
|
scaffold attachment factor B |
| chr16_+_26612401 | 0.07 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr3_+_52684556 | 0.06 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr21_+_27189490 | 0.06 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr17_-_24364527 | 0.06 |
ENSDART00000155192
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr8_+_2530065 | 0.06 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr9_-_10068004 | 0.06 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr11_+_5666325 | 0.06 |
ENSDART00000092270
|
r3hdm4
|
R3H domain containing 4 |
| chr3_-_16039619 | 0.05 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_22313533 | 0.05 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr10_-_7555660 | 0.05 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr1_+_55752593 | 0.05 |
ENSDART00000108838
ENSDART00000134770 |
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr19_-_10295173 | 0.05 |
ENSDART00000136697
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr8_-_2602572 | 0.04 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr12_+_9542124 | 0.03 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr3_+_34985895 | 0.02 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr23_-_27442544 | 0.01 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr24_-_23716097 | 0.01 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr10_+_15454745 | 0.00 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1d2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 1.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 1.5 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 0.6 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 0.6 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 1.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.6 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.3 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.3 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.5 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 1.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 1.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.4 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.3 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.7 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.1 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 1.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.1 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.9 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |