Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
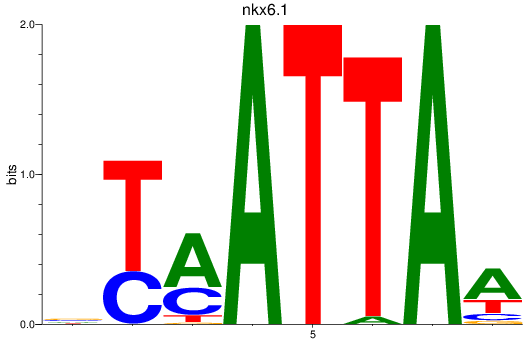
Results for nkx6.1
Z-value: 0.97
Transcription factors associated with nkx6.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.1
|
ENSDARG00000022569 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.1 | dr11_v1_chr21_+_19070921_19070921 | 0.98 | 4.5e-03 | Click! |
Activity profile of nkx6.1 motif
Sorted Z-values of nkx6.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_46111849 | 1.14 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_-_50781623 | 1.04 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr13_+_38430466 | 1.02 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr23_+_40460333 | 0.99 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr9_-_32753535 | 0.97 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr17_-_16965809 | 0.91 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr17_+_23298928 | 0.80 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr10_+_22381802 | 0.79 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr5_+_20147830 | 0.77 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr12_+_24342303 | 0.76 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr7_+_25059845 | 0.76 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr16_+_5774977 | 0.76 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr5_-_23362602 | 0.76 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr14_+_50770537 | 0.75 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr17_+_15433518 | 0.74 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_+_31282161 | 0.74 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr17_-_29119362 | 0.74 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr17_+_15433671 | 0.74 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_-_43538328 | 0.72 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr17_-_12389259 | 0.68 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr21_+_13366353 | 0.68 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr23_+_28731379 | 0.68 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr23_+_20563779 | 0.64 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr15_-_16098531 | 0.64 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr2_+_50608099 | 0.64 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr4_+_21129752 | 0.62 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr8_-_25120231 | 0.61 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr1_+_34181581 | 0.60 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr22_-_13851297 | 0.60 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr8_+_24861264 | 0.60 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr13_+_38521152 | 0.59 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_-_45042210 | 0.59 |
ENSDART00000073694
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr10_-_24371312 | 0.59 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr7_-_28148310 | 0.59 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr1_+_12767318 | 0.59 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr14_-_4556896 | 0.58 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr1_+_12766351 | 0.58 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr8_+_7144066 | 0.58 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr2_+_20332044 | 0.57 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr19_-_31522625 | 0.57 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr16_+_34531486 | 0.57 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr18_+_9637744 | 0.57 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr5_+_36611128 | 0.56 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr8_-_19051906 | 0.56 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr15_-_34213898 | 0.56 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr5_+_63668735 | 0.56 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr9_-_20372977 | 0.56 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_+_8601935 | 0.55 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr12_+_5081759 | 0.54 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr20_+_18551657 | 0.53 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr24_+_2519761 | 0.53 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr9_-_31278048 | 0.52 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr15_+_22311803 | 0.52 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr24_-_29963858 | 0.52 |
ENSDART00000183442
|
CR352310.1
|
|
| chr1_+_33969015 | 0.52 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr25_-_20378721 | 0.51 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr11_-_37509001 | 0.51 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr4_+_12615836 | 0.51 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr14_+_14662116 | 0.51 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr20_+_41756996 | 0.51 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr18_-_1185772 | 0.51 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr7_+_19552381 | 0.51 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr19_-_32641725 | 0.51 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr21_-_42100471 | 0.50 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr6_-_41229787 | 0.50 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr20_+_31076488 | 0.50 |
ENSDART00000136255
ENSDART00000008840 |
otofa
|
otoferlin a |
| chr14_+_5936996 | 0.50 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr5_+_19314574 | 0.50 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr3_-_23406964 | 0.50 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr10_+_29698467 | 0.49 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr21_+_6780340 | 0.49 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr16_+_20161805 | 0.49 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr6_-_10320676 | 0.48 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr12_-_35787801 | 0.48 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr20_-_27711970 | 0.48 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr7_-_28147838 | 0.48 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr3_-_40054615 | 0.48 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr3_-_18711288 | 0.47 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr19_+_6938289 | 0.46 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr20_-_45060241 | 0.46 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr1_-_44701313 | 0.46 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr23_-_26522760 | 0.46 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr20_+_40457599 | 0.46 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr18_+_34478959 | 0.45 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr21_+_28958471 | 0.45 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr20_-_28800999 | 0.45 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr2_-_40135942 | 0.45 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr6_-_38419318 | 0.45 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr8_-_34052019 | 0.45 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr22_-_11136625 | 0.45 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr3_+_34919810 | 0.45 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr8_-_42238543 | 0.44 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr9_-_51436377 | 0.44 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr12_+_35654749 | 0.43 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr3_+_29714775 | 0.43 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr22_-_7129631 | 0.42 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr12_-_26383242 | 0.42 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr11_+_36158134 | 0.42 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr14_+_49135264 | 0.42 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr4_-_8903240 | 0.41 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr12_-_33972798 | 0.41 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr9_-_3671911 | 0.41 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr19_-_31402429 | 0.41 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr5_-_31926906 | 0.41 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr1_-_22512063 | 0.40 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr2_+_38554260 | 0.40 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr6_+_39232245 | 0.40 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr10_+_37137464 | 0.40 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr19_-_5103313 | 0.40 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr25_+_15273370 | 0.40 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr18_+_24921587 | 0.40 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr15_-_6247775 | 0.39 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr8_-_39952727 | 0.39 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr3_+_33341640 | 0.39 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr6_-_35779348 | 0.38 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr3_-_28258462 | 0.38 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr24_+_5208171 | 0.38 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr3_-_35800221 | 0.38 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr10_-_34871737 | 0.38 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_50888806 | 0.38 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr1_+_25801648 | 0.37 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr14_+_45406299 | 0.37 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr11_-_8126223 | 0.37 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr9_+_24159280 | 0.37 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr25_-_13842618 | 0.37 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr7_-_69521481 | 0.36 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr9_+_34641237 | 0.36 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr11_+_30057762 | 0.35 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr12_+_18681477 | 0.35 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr5_-_25174420 | 0.35 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr11_+_36231248 | 0.35 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr17_-_19019635 | 0.35 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_32683089 | 0.35 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr10_+_42423318 | 0.35 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr25_-_1720736 | 0.35 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr1_-_19215336 | 0.34 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr2_+_51183320 | 0.34 |
ENSDART00000167430
|
lrrc24
|
leucine rich repeat containing 24 |
| chr18_-_42333428 | 0.34 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr6_-_44044385 | 0.33 |
ENSDART00000075497
|
rybpb
|
RING1 and YY1 binding protein b |
| chr19_+_5480327 | 0.33 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr1_-_50859053 | 0.33 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr10_+_41765944 | 0.33 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr5_-_16996482 | 0.32 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr18_+_10884996 | 0.32 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr14_-_2933185 | 0.32 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr9_+_17983463 | 0.32 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr11_+_28476298 | 0.32 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr7_-_24699985 | 0.32 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr3_-_20040636 | 0.32 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr1_-_36152131 | 0.32 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr10_-_34870667 | 0.32 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr20_-_29864390 | 0.32 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr2_-_30668580 | 0.32 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr1_+_11977426 | 0.32 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr6_-_12172424 | 0.32 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr23_+_28648864 | 0.32 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr19_+_21362553 | 0.32 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr13_-_30700460 | 0.31 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr7_+_31145386 | 0.31 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr9_+_24159725 | 0.31 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr16_+_17389116 | 0.31 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr20_-_8304271 | 0.30 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr12_+_32073660 | 0.30 |
ENSDART00000153245
ENSDART00000153268 |
stxbp4
|
syntaxin binding protein 4 |
| chr15_-_20024205 | 0.30 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr21_-_18648861 | 0.30 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr10_+_41765616 | 0.30 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr16_-_17162843 | 0.30 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr3_-_34337969 | 0.30 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr11_-_12998400 | 0.30 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr4_+_12612145 | 0.30 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr11_+_23760470 | 0.30 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr1_+_52929185 | 0.30 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr20_-_39271844 | 0.29 |
ENSDART00000192708
|
clu
|
clusterin |
| chr1_-_20271138 | 0.29 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr5_-_34185115 | 0.29 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr21_-_39177564 | 0.29 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr9_-_1702648 | 0.29 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr20_-_20930926 | 0.29 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr19_+_2279051 | 0.28 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr6_-_46162585 | 0.28 |
ENSDART00000017309
|
ca16b
|
carbonic anhydrase XVI b |
| chr18_-_2433011 | 0.28 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr16_+_2820340 | 0.28 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr11_+_24620742 | 0.28 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr12_-_28881638 | 0.28 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr15_-_22074315 | 0.28 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr6_+_21001264 | 0.27 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr11_+_30000814 | 0.27 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr14_-_4273396 | 0.27 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr17_-_37052622 | 0.27 |
ENSDART00000186408
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr20_-_20931197 | 0.27 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr22_-_3564563 | 0.27 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr4_+_5798223 | 0.26 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr23_+_6795709 | 0.26 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr9_+_33009284 | 0.26 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr7_-_54217547 | 0.26 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr7_-_24994722 | 0.26 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr23_+_20705849 | 0.26 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr19_-_7358184 | 0.26 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr5_+_45976130 | 0.26 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr13_+_36622100 | 0.26 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 1.0 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.2 | 1.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.5 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 1.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.5 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.8 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.6 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.6 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.3 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.2 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.2 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.2 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.2 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.3 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.3 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 1.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.6 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.4 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.6 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.8 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.0 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.0 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.6 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.6 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.1 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.2 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 2.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.2 | 0.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.5 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.2 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 0.6 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.0 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.2 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.8 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.5 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |