Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
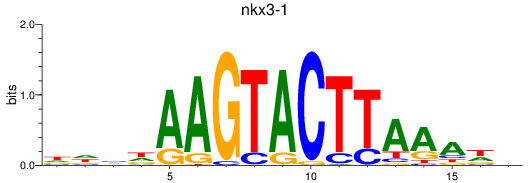
Results for nkx3-1
Z-value: 1.51
Transcription factors associated with nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-1
|
ENSDARG00000078280 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3-1 | dr11_v1_chr8_-_50259448_50259448 | 0.16 | 8.0e-01 | Click! |
Activity profile of nkx3-1 motif
Sorted Z-values of nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25249193 | 1.15 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr22_+_25236657 | 1.12 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr9_+_8396755 | 0.98 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr11_-_30636163 | 0.94 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr2_+_11031360 | 0.80 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr1_-_5542565 | 0.78 |
ENSDART00000160791
ENSDART00000103754 |
fn1b
|
fibronectin 1b |
| chr22_-_11623063 | 0.76 |
ENSDART00000145653
|
gcga
|
glucagon a |
| chr2_+_4208323 | 0.75 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr19_-_5699703 | 0.74 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr2_+_1486822 | 0.74 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr17_+_24615091 | 0.73 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| chr6_-_54826061 | 0.73 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr8_-_40464935 | 0.73 |
ENSDART00000040013
|
myl7
|
myosin, light chain 7, regulatory |
| chr9_+_44994214 | 0.72 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr2_+_1487118 | 0.71 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr25_+_31323978 | 0.70 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr11_-_5865744 | 0.68 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr16_+_54641230 | 0.68 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr8_-_39739627 | 0.64 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr22_+_21324398 | 0.62 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr3_+_29942338 | 0.61 |
ENSDART00000158220
|
ifi35
|
interferon-induced protein 35 |
| chr4_-_9891874 | 0.58 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr23_+_31431956 | 0.56 |
ENSDART00000138350
ENSDART00000053542 |
rps12
|
ribosomal protein S12 |
| chr14_+_52440161 | 0.55 |
ENSDART00000168437
|
b4galt1l
|
DP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1, like |
| chr8_+_6576940 | 0.55 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr25_-_13403726 | 0.54 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr22_+_12770877 | 0.53 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr1_-_59169815 | 0.52 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr23_-_10254288 | 0.52 |
ENSDART00000081215
|
krt8
|
keratin 8 |
| chr20_-_33566640 | 0.52 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr8_-_39739056 | 0.52 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr16_-_31686602 | 0.51 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr2_-_879800 | 0.51 |
ENSDART00000019733
|
irf4a
|
interferon regulatory factor 4a |
| chr7_+_8358547 | 0.50 |
ENSDART00000173016
|
jac7
|
jacalin 7 |
| chr15_+_29025090 | 0.49 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_-_33507458 | 0.47 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr24_-_30263301 | 0.47 |
ENSDART00000162328
|
snx7
|
sorting nexin 7 |
| chr3_+_22377312 | 0.47 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr5_-_64883082 | 0.46 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr15_-_28805493 | 0.46 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr4_+_76671012 | 0.45 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr14_+_48903640 | 0.45 |
ENSDART00000165323
|
zgc:154054
|
zgc:154054 |
| chr16_+_31922065 | 0.45 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr23_+_25832689 | 0.44 |
ENSDART00000138907
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr9_-_12888082 | 0.43 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr5_-_25037454 | 0.43 |
ENSDART00000027237
|
abo
|
ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) |
| chr16_-_51299061 | 0.43 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr24_-_27473771 | 0.43 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr19_-_42556086 | 0.42 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr19_-_31372896 | 0.42 |
ENSDART00000046609
|
scin
|
scinderin |
| chr6_-_11091449 | 0.42 |
ENSDART00000127209
|
pttg1ipa
|
PTTG1 interacting protein a |
| chr7_+_17947217 | 0.42 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr13_+_159504 | 0.42 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr2_-_10877228 | 0.42 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr7_+_2455344 | 0.41 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr18_+_40471826 | 0.41 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr13_-_37647209 | 0.41 |
ENSDART00000189102
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr11_-_40128722 | 0.41 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr5_-_67499279 | 0.40 |
ENSDART00000128050
|
si:dkey-251i10.3
|
si:dkey-251i10.3 |
| chr18_+_6558338 | 0.40 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr9_-_51563575 | 0.40 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr21_-_30658509 | 0.40 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr4_-_4119396 | 0.40 |
ENSDART00000067409
ENSDART00000138221 |
lmod2b
|
leiomodin 2 (cardiac) b |
| chr16_+_33142734 | 0.39 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr12_-_13155653 | 0.39 |
ENSDART00000152467
|
cxl34c
|
CX chemokine ligand 34c |
| chr15_-_39969988 | 0.39 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr5_-_10082244 | 0.39 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr19_+_4051695 | 0.38 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr3_+_29941777 | 0.38 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr16_-_4203022 | 0.37 |
ENSDART00000018686
|
rrp15
|
ribosomal RNA processing 15 homolog |
| chr10_-_34915886 | 0.37 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr3_-_13921173 | 0.37 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr10_-_34916208 | 0.37 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr24_-_26328721 | 0.37 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr16_+_24842662 | 0.37 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr12_-_5418340 | 0.36 |
ENSDART00000028043
|
noc3l
|
NOC3-like DNA replication regulator |
| chr5_+_26795773 | 0.36 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr11_+_20899029 | 0.36 |
ENSDART00000163029
|
zgc:162182
|
zgc:162182 |
| chr11_+_24729346 | 0.35 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr4_+_76735113 | 0.35 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr8_-_38105053 | 0.35 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr7_+_12950507 | 0.34 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr24_+_26328787 | 0.34 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr9_-_50001606 | 0.33 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr4_+_76705830 | 0.33 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr3_+_25849560 | 0.33 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr17_-_1705013 | 0.32 |
ENSDART00000182864
|
CABZ01086293.1
|
|
| chr10_-_3427589 | 0.32 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr5_+_36666715 | 0.32 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr25_-_13050959 | 0.32 |
ENSDART00000169041
|
ccl35.1
|
chemokine (C-C motif) ligand 35, duplicate 1 |
| chr14_+_14806851 | 0.32 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr12_+_23850661 | 0.31 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr17_+_30894431 | 0.31 |
ENSDART00000127996
|
degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr17_-_6765877 | 0.30 |
ENSDART00000193102
|
CR352340.1
|
|
| chr2_-_7757273 | 0.30 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr20_+_25552057 | 0.29 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr21_+_25198637 | 0.29 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr19_+_7424347 | 0.29 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr12_-_28548789 | 0.29 |
ENSDART00000153062
|
si:ch73-180n10.1
|
si:ch73-180n10.1 |
| chr18_-_22753637 | 0.28 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr20_-_25902141 | 0.28 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr17_+_6538733 | 0.28 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr13_+_27328098 | 0.28 |
ENSDART00000037585
|
mb21d1
|
Mab-21 domain containing 1 |
| chr7_-_20464468 | 0.28 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr23_+_4362463 | 0.28 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr6_+_7466223 | 0.28 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr22_+_13886821 | 0.27 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr7_+_41322407 | 0.27 |
ENSDART00000114076
ENSDART00000139093 |
dph2
|
DPH2 homolog (S. cerevisiae) |
| chr9_-_8296723 | 0.27 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr8_-_38317914 | 0.27 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr9_+_35015747 | 0.27 |
ENSDART00000140110
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr1_+_24365582 | 0.27 |
ENSDART00000169389
|
pimr178
|
Pim proto-oncogene, serine/threonine kinase, related 178 |
| chr3_+_3139240 | 0.26 |
ENSDART00000105014
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr3_+_16922226 | 0.26 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr14_+_8715527 | 0.25 |
ENSDART00000170754
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr13_-_48058779 | 0.25 |
ENSDART00000186623
ENSDART00000045475 |
itga9
|
integrin, alpha 9 |
| chr25_+_10953351 | 0.25 |
ENSDART00000154050
|
mhc1lda
|
major histocompatibility complex class I LDA |
| chr5_+_1933131 | 0.25 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr12_-_30548244 | 0.25 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr8_+_30784020 | 0.25 |
ENSDART00000112981
|
tas2r200.1
|
taste receptor, type 2, member 200, tandem duplicate 1 |
| chr1_-_55248496 | 0.25 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr18_+_35861930 | 0.25 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr16_-_9425451 | 0.24 |
ENSDART00000149163
|
ccr8.1
|
chemokine (C-C motif) receptor 8.1 |
| chr2_+_10771787 | 0.24 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr21_+_19547806 | 0.24 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr15_+_25452092 | 0.24 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr12_-_31461219 | 0.24 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr5_+_27429872 | 0.24 |
ENSDART00000087894
|
zgc:165555
|
zgc:165555 |
| chr7_+_2259413 | 0.23 |
ENSDART00000173420
|
si:dkey-187j14.7
|
si:dkey-187j14.7 |
| chr3_-_20957011 | 0.23 |
ENSDART00000159787
ENSDART00000180531 |
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr20_+_33904258 | 0.23 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr19_-_10330778 | 0.23 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr21_+_10828828 | 0.23 |
ENSDART00000136449
|
oacyl
|
O-acyltransferase like |
| chr18_-_6766354 | 0.23 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr7_+_13702248 | 0.22 |
ENSDART00000172811
|
pdcd7
|
programmed cell death 7 |
| chr21_+_25236297 | 0.22 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr7_+_22981441 | 0.22 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr2_-_11101329 | 0.22 |
ENSDART00000164416
|
lrrc53
|
leucine rich repeat containing 53 |
| chr5_-_16475682 | 0.22 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_-_57204352 | 0.22 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr4_-_67800414 | 0.22 |
ENSDART00000160213
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr7_+_22680560 | 0.21 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr9_+_15893093 | 0.21 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr24_-_31904924 | 0.21 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr4_-_18434924 | 0.20 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr5_-_23795688 | 0.20 |
ENSDART00000099084
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr16_-_42175617 | 0.20 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr9_-_52386733 | 0.20 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr16_-_41535690 | 0.20 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr1_+_57040472 | 0.20 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr19_+_45962016 | 0.19 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr15_+_38025319 | 0.19 |
ENSDART00000155000
|
si:ch73-380l3.3
|
si:ch73-380l3.3 |
| chr15_+_12030367 | 0.19 |
ENSDART00000176178
|
FO904901.1
|
|
| chr20_-_18535502 | 0.18 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_-_354115 | 0.18 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr17_-_43623356 | 0.17 |
ENSDART00000075596
ENSDART00000136431 |
rtkn2a
|
rhotekin 2a |
| chr7_+_48667081 | 0.17 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr14_+_14806692 | 0.17 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr4_-_65323874 | 0.17 |
ENSDART00000175271
ENSDART00000187913 |
BX571813.1
|
|
| chr18_+_2228737 | 0.17 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr15_+_17848590 | 0.17 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr1_-_5455498 | 0.17 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr13_-_31370184 | 0.17 |
ENSDART00000034829
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr22_-_11626014 | 0.16 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr1_-_58002973 | 0.16 |
ENSDART00000140390
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr12_+_33361948 | 0.16 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr6_+_11438972 | 0.16 |
ENSDART00000029314
|
col5a2b
|
collagen, type V, alpha 2b |
| chr13_+_7242916 | 0.16 |
ENSDART00000184238
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr12_-_4424424 | 0.16 |
ENSDART00000152314
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr9_+_35016201 | 0.16 |
ENSDART00000182404
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr14_+_3495542 | 0.15 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr19_-_18152942 | 0.15 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr12_+_17100021 | 0.15 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr8_+_25616946 | 0.15 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr24_+_24086491 | 0.15 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr22_-_23000815 | 0.14 |
ENSDART00000137111
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr8_+_45004997 | 0.14 |
ENSDART00000159779
|
si:ch211-163b2.4
|
si:ch211-163b2.4 |
| chr23_+_36340520 | 0.14 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr5_-_16475374 | 0.14 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr21_-_11286483 | 0.14 |
ENSDART00000074845
|
rtkn2b
|
rhotekin 2b |
| chr13_-_41947996 | 0.14 |
ENSDART00000132808
|
nanp
|
N-acetylneuraminic acid phosphatase |
| chr4_+_3312852 | 0.14 |
ENSDART00000084691
|
fgd4b
|
FYVE, RhoGEF and PH domain containing 4b |
| chr1_-_2457546 | 0.14 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr21_+_944715 | 0.14 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr6_+_72040 | 0.13 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr17_-_24703778 | 0.13 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr10_-_8957480 | 0.13 |
ENSDART00000150049
|
mocs2
|
molybdenum cofactor synthesis 2 |
| chr2_+_33747509 | 0.13 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr17_+_14886828 | 0.13 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr15_+_37331585 | 0.13 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr23_+_35672542 | 0.13 |
ENSDART00000046268
|
pmelb
|
premelanosome protein b |
| chr14_-_48786708 | 0.13 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr3_-_13599482 | 0.13 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr17_-_43031763 | 0.13 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr4_+_76745015 | 0.13 |
ENSDART00000155883
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr3_-_22366562 | 0.13 |
ENSDART00000129447
|
ifnphi1
|
interferon phi 1 |
| chr4_+_54948890 | 0.12 |
ENSDART00000166873
|
si:dkey-56m15.7
|
si:dkey-56m15.7 |
| chr10_+_15310811 | 0.12 |
ENSDART00000136239
|
kcnv2a
|
potassium channel, subfamily V, member 2a |
| chr2_-_10641998 | 0.12 |
ENSDART00000188323
|
mtf2
|
metal response element binding transcription factor 2 |
| chr16_+_11603096 | 0.12 |
ENSDART00000146286
|
si:dkey-11o1.3
|
si:dkey-11o1.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.5 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.2 | 0.7 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.5 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.2 | 1.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.7 | GO:0055014 | ventricular cardiac myofibril assembly(GO:0055005) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.3 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.6 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.7 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.5 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.3 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 1.4 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.5 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0034311 | diol metabolic process(GO:0034311) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0090133 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.8 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.2 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |