Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
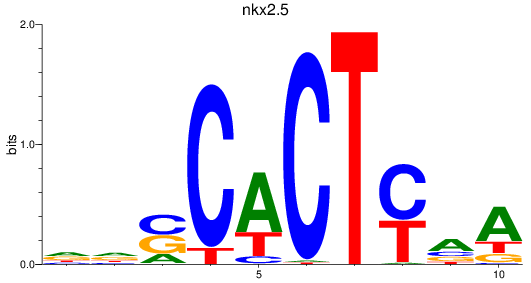
Results for nkx2.5
Z-value: 0.74
Transcription factors associated with nkx2.5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.5
|
ENSDARG00000018004 | NK2 homeobox 5 |
|
nkx2.5
|
ENSDARG00000116714 | NK2 homeobox 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.5 | dr11_v1_chr14_+_24241241_24241241 | 0.56 | 3.3e-01 | Click! |
Activity profile of nkx2.5 motif
Sorted Z-values of nkx2.5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_30475563 | 0.30 |
ENSDART00000133118
|
gja5a
|
gap junction protein, alpha 5a |
| chr8_+_48609521 | 0.29 |
ENSDART00000060765
|
nppb
|
natriuretic peptide B |
| chr7_-_16562200 | 0.27 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr4_-_4119396 | 0.25 |
ENSDART00000067409
ENSDART00000138221 |
lmod2b
|
leiomodin 2 (cardiac) b |
| chr12_-_26423439 | 0.22 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr13_+_28702104 | 0.22 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr10_+_1052591 | 0.21 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr20_-_28642061 | 0.20 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr3_+_19207176 | 0.20 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr25_-_28674739 | 0.20 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr2_+_55916911 | 0.19 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr15_+_44366556 | 0.18 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr3_+_14463941 | 0.18 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr22_+_25236888 | 0.17 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr18_-_50986704 | 0.16 |
ENSDART00000191435
|
LO018598.1
|
|
| chr21_+_31434251 | 0.15 |
ENSDART00000040740
ENSDART00000130157 |
si:ch211-12m10.1
si:ch211-166i24.1
|
si:ch211-12m10.1 si:ch211-166i24.1 |
| chr20_-_28698172 | 0.15 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr9_+_2452672 | 0.15 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr3_+_54744069 | 0.15 |
ENSDART00000134958
ENSDART00000114443 |
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr23_-_333457 | 0.14 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr3_+_22905341 | 0.14 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr19_-_28360033 | 0.14 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr1_-_46981134 | 0.13 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr20_+_42918755 | 0.13 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chrM_+_4993 | 0.13 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr23_+_2778813 | 0.13 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr22_+_25242322 | 0.13 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr23_-_24856025 | 0.13 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr18_+_17534627 | 0.13 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr22_+_724639 | 0.13 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr22_+_21317597 | 0.13 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr1_+_50538839 | 0.13 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr6_+_24817852 | 0.13 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr21_-_25484990 | 0.12 |
ENSDART00000180289
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr3_+_56163276 | 0.12 |
ENSDART00000166709
ENSDART00000158540 |
PRKCA
|
si:ch73-374l24.1 |
| chr19_-_2115040 | 0.12 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr16_+_34493987 | 0.12 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr22_+_1421212 | 0.12 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr1_+_34243650 | 0.12 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr16_+_38240027 | 0.12 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr11_-_29082175 | 0.12 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr5_-_29122615 | 0.11 |
ENSDART00000144802
|
whrnb
|
whirlin b |
| chr17_-_608857 | 0.11 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr11_-_29082429 | 0.11 |
ENSDART00000041443
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr3_-_10582384 | 0.11 |
ENSDART00000048095
ENSDART00000155152 |
elac2
|
elaC ribonuclease Z 2 |
| chr5_+_51111766 | 0.11 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr2_+_30513887 | 0.11 |
ENSDART00000137048
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr2_-_17827983 | 0.10 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr8_+_3431671 | 0.10 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr8_+_7756893 | 0.10 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_+_37424261 | 0.10 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr20_-_42102416 | 0.10 |
ENSDART00000186378
ENSDART00000188253 ENSDART00000186458 |
slc35f1
|
solute carrier family 35, member F1 |
| chr24_-_11057305 | 0.10 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr10_-_24371312 | 0.10 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr17_-_44249538 | 0.10 |
ENSDART00000008816
|
otx2b
|
orthodenticle homeobox 2b |
| chr19_-_6873107 | 0.10 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr11_-_27537593 | 0.10 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr5_+_1983495 | 0.10 |
ENSDART00000179860
|
CT573103.2
|
|
| chr3_-_5664123 | 0.10 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr4_-_4795205 | 0.10 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr6_-_10168822 | 0.09 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr18_-_47662696 | 0.09 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr16_-_12097558 | 0.09 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr12_+_26706745 | 0.09 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr20_+_50956369 | 0.09 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr13_-_50139916 | 0.09 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr20_+_25568694 | 0.09 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr17_+_30442518 | 0.09 |
ENSDART00000155264
|
lpin1
|
lipin 1 |
| chr1_+_46598764 | 0.09 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr3_+_40856095 | 0.09 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr5_+_47975758 | 0.09 |
ENSDART00000097429
|
BX470189.1
|
|
| chr3_+_34919810 | 0.08 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr22_-_30881738 | 0.08 |
ENSDART00000059965
|
si:dkey-49n23.1
|
si:dkey-49n23.1 |
| chr8_+_25254435 | 0.08 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr17_-_16342388 | 0.08 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr6_-_1820606 | 0.08 |
ENSDART00000183228
|
FO834857.1
|
|
| chr24_-_23839647 | 0.08 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr12_+_32292564 | 0.08 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr10_-_38498072 | 0.08 |
ENSDART00000111234
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr22_+_25236657 | 0.08 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr2_+_269781 | 0.08 |
ENSDART00000156019
|
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr9_+_17862858 | 0.08 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr20_+_2460864 | 0.08 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr2_+_27010439 | 0.08 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr7_-_23777445 | 0.08 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr20_-_25671342 | 0.08 |
ENSDART00000182775
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr12_+_44610912 | 0.08 |
ENSDART00000179030
ENSDART00000178008 |
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr15_-_21132480 | 0.08 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr14_+_16036139 | 0.07 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr1_-_14694809 | 0.07 |
ENSDART00000193331
|
CU138548.5
|
|
| chr22_+_2830703 | 0.07 |
ENSDART00000145463
ENSDART00000144785 |
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr1_+_46598502 | 0.07 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr10_-_26218354 | 0.07 |
ENSDART00000180764
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr18_-_399554 | 0.07 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr17_+_25955003 | 0.07 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr15_-_47937204 | 0.07 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr3_-_13599482 | 0.07 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr11_-_34232906 | 0.07 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr6_-_7726849 | 0.07 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr7_-_32833153 | 0.07 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr12_-_1694053 | 0.07 |
ENSDART00000169370
|
CABZ01081830.2
|
|
| chr23_-_14769523 | 0.07 |
ENSDART00000054909
|
gss
|
glutathione synthetase |
| chr6_+_515181 | 0.07 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr17_-_44249720 | 0.07 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr11_+_37216668 | 0.07 |
ENSDART00000173076
|
zgc:112265
|
zgc:112265 |
| chr23_+_7710447 | 0.07 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr22_+_1481307 | 0.07 |
ENSDART00000177429
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr12_+_2870671 | 0.06 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr3_+_24459709 | 0.06 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr3_+_39600562 | 0.06 |
ENSDART00000134309
ENSDART00000007170 |
prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr13_-_8229977 | 0.06 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr2_-_37280028 | 0.06 |
ENSDART00000139459
|
nadkb
|
NAD kinase b |
| chr8_-_4100365 | 0.06 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr23_+_7710721 | 0.06 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr17_+_23146976 | 0.06 |
ENSDART00000114212
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr7_-_40122139 | 0.06 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr10_+_8481660 | 0.06 |
ENSDART00000109559
|
BX682234.1
|
|
| chr14_-_2267515 | 0.06 |
ENSDART00000180988
ENSDART00000130712 |
pcdh2aa15
|
protocadherin 2 alpha a 15 |
| chr5_-_29122834 | 0.06 |
ENSDART00000087197
|
whrnb
|
whirlin b |
| chr23_+_30736895 | 0.06 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr14_+_11762991 | 0.06 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_-_43850637 | 0.06 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr22_+_38159823 | 0.06 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr7_-_35083585 | 0.06 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr14_+_31493306 | 0.06 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr6_+_11250316 | 0.05 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr22_+_18389271 | 0.05 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr22_+_28818291 | 0.05 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr15_-_21155641 | 0.05 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr22_+_39058269 | 0.05 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr17_-_45155181 | 0.05 |
ENSDART00000185274
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr5_+_24543862 | 0.05 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr10_-_34867401 | 0.05 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr24_+_15655233 | 0.05 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr5_-_62317496 | 0.05 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr11_-_8782871 | 0.05 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr4_-_77125693 | 0.05 |
ENSDART00000174256
|
CU467646.3
|
|
| chr17_-_7436766 | 0.05 |
ENSDART00000162002
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr17_+_30205258 | 0.05 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr3_-_7170661 | 0.05 |
ENSDART00000190345
|
BX005085.4
|
|
| chr6_-_18121075 | 0.05 |
ENSDART00000171072
|
SEC14L1
|
si:dkey-237i9.1 |
| chr22_+_1330477 | 0.05 |
ENSDART00000157567
|
CU207221.2
|
|
| chr15_-_26568278 | 0.05 |
ENSDART00000182609
|
wdr81
|
WD repeat domain 81 |
| chr18_-_29977431 | 0.05 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr1_+_16621345 | 0.05 |
ENSDART00000149026
|
pcm1
|
pericentriolar material 1 |
| chr22_+_1480988 | 0.04 |
ENSDART00000171122
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr6_-_12900154 | 0.04 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr12_-_19862912 | 0.04 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr6_-_42036685 | 0.04 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr20_+_44311448 | 0.04 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr14_+_31493119 | 0.04 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr5_+_31944270 | 0.04 |
ENSDART00000147850
|
ungb
|
uracil DNA glycosylase b |
| chr9_-_13871935 | 0.04 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr4_-_77130289 | 0.04 |
ENSDART00000174380
|
CU467646.7
|
|
| chr14_+_23874062 | 0.04 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr2_-_37280617 | 0.04 |
ENSDART00000190458
|
nadkb
|
NAD kinase b |
| chr9_+_37320222 | 0.04 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr16_-_17200120 | 0.04 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr13_-_28610965 | 0.04 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr14_-_16476863 | 0.04 |
ENSDART00000089021
|
canx
|
calnexin |
| chr16_-_41787421 | 0.04 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr10_-_29831944 | 0.04 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr14_-_32255126 | 0.04 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr25_+_36152215 | 0.04 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr23_-_6879731 | 0.04 |
ENSDART00000082154
|
trim35-39
|
tripartite motif containing 35-39 |
| chr19_+_26423590 | 0.04 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr11_-_3629201 | 0.04 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr13_-_6218248 | 0.04 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr6_-_6673813 | 0.04 |
ENSDART00000150995
|
si:dkey-261j15.2
|
si:dkey-261j15.2 |
| chr24_+_10039165 | 0.04 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr2_-_40890004 | 0.04 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr3_+_48234445 | 0.04 |
ENSDART00000161419
|
tbcd
|
tubulin folding cofactor D |
| chr1_+_27024068 | 0.03 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr6_-_42290259 | 0.03 |
ENSDART00000064969
|
tex264a
|
testis expressed 264a |
| chr21_+_27513859 | 0.03 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr15_-_11955485 | 0.03 |
ENSDART00000160286
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr2_-_40890264 | 0.03 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr5_+_26765275 | 0.03 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr5_-_64511428 | 0.03 |
ENSDART00000016321
|
rxrab
|
retinoid x receptor, alpha b |
| chr1_+_27023687 | 0.03 |
ENSDART00000189093
|
bnc2
|
basonuclin 2 |
| chr22_+_36914636 | 0.03 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr23_+_26946429 | 0.03 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr7_-_30560400 | 0.03 |
ENSDART00000142680
ENSDART00000142818 |
sltm
|
SAFB-like, transcription modulator |
| chr18_+_41232719 | 0.03 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr13_-_32899322 | 0.03 |
ENSDART00000133882
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr5_-_31712399 | 0.03 |
ENSDART00000141328
|
pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr24_-_26945390 | 0.03 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr24_+_26017094 | 0.03 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr4_-_77135076 | 0.03 |
ENSDART00000174184
|
zgc:173770
|
zgc:173770 |
| chr19_+_7810028 | 0.03 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr19_+_7292654 | 0.03 |
ENSDART00000140459
|
ccdc127b
|
coiled-coil domain containing 127b |
| chr15_+_24212847 | 0.03 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr12_-_45304971 | 0.03 |
ENSDART00000186537
ENSDART00000126405 |
fdxr
|
ferredoxin reductase |
| chr21_+_13205859 | 0.03 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr1_-_20911297 | 0.03 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr8_+_21114338 | 0.03 |
ENSDART00000002186
|
uck2a
|
uridine-cytidine kinase 2a |
| chr19_-_29832876 | 0.03 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr19_+_7292445 | 0.03 |
ENSDART00000026634
|
ccdc127b
|
coiled-coil domain containing 127b |
| chr24_-_7587401 | 0.03 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr5_+_12515402 | 0.03 |
ENSDART00000103278
|
zgc:171242
|
zgc:171242 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.3 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.0 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |