Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for nkx2.2a
Z-value: 0.78
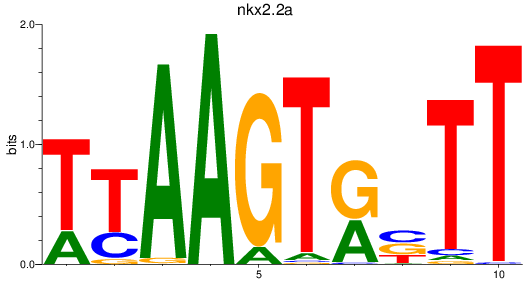
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr11_v1_chr17_-_42218652_42218652 | 0.16 | 8.0e-01 | Click! |
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_28349144 | 0.55 |
ENSDART00000179690
ENSDART00000188059 |
ino80
|
INO80 complex subunit |
| chr22_+_21317597 | 0.38 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr25_-_32311048 | 0.38 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr5_+_1983495 | 0.38 |
ENSDART00000179860
|
CT573103.2
|
|
| chr24_+_22039964 | 0.37 |
ENSDART00000081220
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr11_-_3629201 | 0.35 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr1_-_45888608 | 0.34 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr23_+_1078072 | 0.33 |
ENSDART00000159263
ENSDART00000053527 |
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr24_-_2829049 | 0.32 |
ENSDART00000164913
|
PPP1R3G
|
si:ch211-152c8.5 |
| chr14_-_42997145 | 0.29 |
ENSDART00000172801
|
pcdh10b
|
protocadherin 10b |
| chr8_-_9118958 | 0.26 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr19_-_32940040 | 0.26 |
ENSDART00000179947
|
azin1b
|
antizyme inhibitor 1b |
| chr22_+_20720808 | 0.26 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr13_-_18637244 | 0.24 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr22_+_25236657 | 0.24 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr19_-_41213718 | 0.24 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr22_+_25249193 | 0.24 |
ENSDART00000171851
|
si:ch211-226h8.11
|
si:ch211-226h8.11 |
| chr5_-_23277939 | 0.23 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr1_+_15137901 | 0.23 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr5_+_72087619 | 0.22 |
ENSDART00000062885
|
oxt
|
oxytocin |
| chr1_+_45323142 | 0.21 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr7_+_41295974 | 0.21 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr8_-_39739627 | 0.21 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr3_-_48603471 | 0.21 |
ENSDART00000189027
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr8_-_1698155 | 0.20 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr1_+_17695426 | 0.20 |
ENSDART00000103236
|
ankrd37
|
ankyrin repeat domain 37 |
| chr7_-_69636502 | 0.20 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr23_+_40460333 | 0.20 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr22_-_8006342 | 0.19 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr25_-_7974494 | 0.19 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr12_-_9438227 | 0.19 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr23_+_27740788 | 0.19 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr20_+_33904258 | 0.19 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr17_-_14726824 | 0.19 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr8_-_39739056 | 0.19 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr22_-_24757785 | 0.18 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr4_-_18436899 | 0.18 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr2_-_20052561 | 0.18 |
ENSDART00000100133
|
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr21_+_9628854 | 0.18 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr9_-_23156908 | 0.18 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr19_+_1184878 | 0.18 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr8_+_28065803 | 0.17 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr13_-_40726865 | 0.17 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr5_+_31965845 | 0.17 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr19_+_47783137 | 0.17 |
ENSDART00000024777
ENSDART00000158979 |
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr2_+_243778 | 0.17 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr23_+_6795709 | 0.16 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr6_+_9793791 | 0.16 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr23_+_27740592 | 0.16 |
ENSDART00000137875
|
dhh
|
desert hedgehog |
| chr20_+_66857 | 0.16 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr19_+_9344171 | 0.16 |
ENSDART00000133447
ENSDART00000104622 |
si:ch211-288g17.4
|
si:ch211-288g17.4 |
| chr12_-_19865585 | 0.16 |
ENSDART00000066386
|
shisa9a
|
shisa family member 9a |
| chr11_+_43114108 | 0.16 |
ENSDART00000013642
ENSDART00000190266 |
foxg1b
|
forkhead box G1b |
| chr13_+_33368140 | 0.15 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr6_+_49255706 | 0.15 |
ENSDART00000156866
|
si:dkey-183k8.2
|
si:dkey-183k8.2 |
| chr4_+_19535946 | 0.15 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr16_-_25400257 | 0.15 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr9_-_18911608 | 0.15 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr2_+_1487118 | 0.15 |
ENSDART00000147283
|
c8a
|
complement component 8, alpha polypeptide |
| chr23_+_6795531 | 0.15 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr19_+_37848830 | 0.15 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr2_+_1486822 | 0.15 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr11_-_7410537 | 0.15 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr17_+_30205258 | 0.15 |
ENSDART00000076596
ENSDART00000153795 |
spata17
|
spermatogenesis associated 17 |
| chr18_-_14937211 | 0.14 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr7_-_26518086 | 0.14 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr8_+_31248917 | 0.14 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr24_-_7699356 | 0.14 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr9_-_51323545 | 0.14 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr13_-_32995324 | 0.14 |
ENSDART00000140542
ENSDART00000037740 |
kcnf1b
|
potassium voltage-gated channel, subfamily F, member 1b |
| chr10_+_9372702 | 0.14 |
ENSDART00000021100
|
lhx6
|
LIM homeobox 6 |
| chr13_+_47710434 | 0.14 |
ENSDART00000188724
|
TMEM87B
|
transmembrane protein 87B |
| chr15_+_19472129 | 0.14 |
ENSDART00000154779
|
zgc:77784
|
zgc:77784 |
| chr17_-_30205302 | 0.14 |
ENSDART00000156827
|
si:dkey-27l15.1
|
si:dkey-27l15.1 |
| chr8_+_51050554 | 0.14 |
ENSDART00000166249
|
DISP3
|
si:dkey-32e23.6 |
| chr5_+_20035284 | 0.14 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr6_+_20650003 | 0.13 |
ENSDART00000049465
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr14_-_26536504 | 0.13 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr2_+_51818039 | 0.13 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr2_+_38924975 | 0.13 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr23_+_36730713 | 0.13 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr23_-_39784368 | 0.13 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr1_+_8534698 | 0.13 |
ENSDART00000021504
|
smcr8b
|
Smith-Magenis syndrome chromosome region, candidate 8b |
| chr24_-_31332087 | 0.13 |
ENSDART00000161179
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr15_-_44601331 | 0.13 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr16_-_41465542 | 0.13 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr9_-_9998087 | 0.13 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr25_+_35375848 | 0.13 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr13_+_33368503 | 0.13 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr12_+_16281312 | 0.13 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr13_+_29352842 | 0.12 |
ENSDART00000113009
|
ogdhl
|
oxoglutarate dehydrogenase like |
| chr3_-_32927516 | 0.12 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr5_+_54685175 | 0.12 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr8_+_3431671 | 0.12 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr1_-_21297748 | 0.12 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr2_+_37424261 | 0.12 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr5_+_16117871 | 0.12 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr7_-_35083585 | 0.12 |
ENSDART00000192732
|
agrp
|
agouti related neuropeptide |
| chr11_+_42422638 | 0.12 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr22_-_17586064 | 0.12 |
ENSDART00000060786
ENSDART00000188303 ENSDART00000181212 ENSDART00000181951 |
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr21_-_25573064 | 0.12 |
ENSDART00000134310
|
CR388166.1
|
|
| chr9_+_21722733 | 0.12 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr6_+_10094061 | 0.12 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr20_-_6812688 | 0.12 |
ENSDART00000170934
|
igfbp1a
|
insulin-like growth factor binding protein 1a |
| chr9_-_22158784 | 0.11 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr9_+_42095220 | 0.11 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr19_-_677713 | 0.11 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr11_-_29623380 | 0.11 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr2_+_22531185 | 0.11 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr1_+_37196106 | 0.11 |
ENSDART00000008756
ENSDART00000157503 ENSDART00000162971 ENSDART00000191004 ENSDART00000078206 ENSDART00000045111 |
dclk2a
|
doublecortin-like kinase 2a |
| chr5_-_58939460 | 0.11 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr15_-_20731638 | 0.11 |
ENSDART00000170616
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr4_-_41269844 | 0.11 |
ENSDART00000186177
|
CR388165.2
|
|
| chr2_-_17044959 | 0.11 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr6_+_1724889 | 0.11 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr23_-_27701361 | 0.11 |
ENSDART00000186688
ENSDART00000183985 |
dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr24_+_5935377 | 0.11 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr13_+_36770738 | 0.11 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr3_+_40856095 | 0.11 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr13_-_29424454 | 0.11 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr10_-_24868581 | 0.11 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr16_+_17763848 | 0.11 |
ENSDART00000149408
ENSDART00000148878 |
them4
|
thioesterase superfamily member 4 |
| chr7_-_38698583 | 0.11 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr14_+_29200772 | 0.11 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr23_-_24856025 | 0.10 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr7_-_41489997 | 0.10 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr21_-_42055872 | 0.10 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr7_-_35083184 | 0.10 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr13_-_24379271 | 0.10 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr2_-_32386942 | 0.10 |
ENSDART00000131408
|
ubtfl
|
upstream binding transcription factor, like |
| chr1_-_354115 | 0.10 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr9_-_31596016 | 0.10 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr25_+_3358701 | 0.10 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr19_-_11157659 | 0.10 |
ENSDART00000146557
|
si:dkey-240h12.3
|
si:dkey-240h12.3 |
| chr8_+_8845932 | 0.10 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr19_+_1835940 | 0.10 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr6_-_10168822 | 0.10 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr6_-_40098641 | 0.09 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr22_-_20924564 | 0.09 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr15_-_13254480 | 0.09 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr23_+_4362463 | 0.09 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr9_+_23900703 | 0.09 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr2_+_20472150 | 0.09 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr2_-_32352946 | 0.09 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr15_-_16070731 | 0.09 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr1_+_36651059 | 0.09 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr5_+_32009080 | 0.09 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr12_-_35883814 | 0.09 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr2_+_7557912 | 0.09 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr5_+_9408901 | 0.09 |
ENSDART00000193364
|
FP236810.1
|
|
| chr16_-_31284922 | 0.09 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr1_+_41690402 | 0.09 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr6_+_21395051 | 0.09 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr16_-_12097394 | 0.09 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr17_+_10501647 | 0.09 |
ENSDART00000140391
|
tyro3
|
TYRO3 protein tyrosine kinase |
| chr21_+_11684830 | 0.09 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr15_-_34322915 | 0.09 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr19_-_22328154 | 0.09 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr1_+_8304904 | 0.09 |
ENSDART00000168631
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr11_+_25064519 | 0.09 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr2_-_38992304 | 0.09 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr16_-_28594181 | 0.09 |
ENSDART00000059053
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr19_+_7552699 | 0.09 |
ENSDART00000180788
ENSDART00000115058 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr16_+_30301539 | 0.09 |
ENSDART00000186018
|
LO017848.1
|
|
| chr11_+_7580079 | 0.09 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr8_+_23485079 | 0.09 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr11_+_18873113 | 0.08 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr13_-_18863680 | 0.08 |
ENSDART00000109277
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr21_+_39948300 | 0.08 |
ENSDART00000137740
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr11_-_3954691 | 0.08 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr21_-_37951819 | 0.08 |
ENSDART00000139549
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr12_-_3237561 | 0.08 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr14_+_3449780 | 0.08 |
ENSDART00000163849
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr12_-_24812403 | 0.08 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr24_+_15655069 | 0.08 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr2_-_11662851 | 0.08 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr23_-_31512496 | 0.08 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr17_+_8324345 | 0.08 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr16_-_28593951 | 0.08 |
ENSDART00000183322
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr1_-_54622227 | 0.08 |
ENSDART00000049010
|
tekt4
|
tektin 4 |
| chr10_-_28395620 | 0.08 |
ENSDART00000168907
|
CR392341.4
|
|
| chr10_+_34394454 | 0.08 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr12_+_36413886 | 0.08 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr12_+_16440708 | 0.08 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr1_-_51261420 | 0.08 |
ENSDART00000168685
|
kif16ba
|
kinesin family member 16Ba |
| chr4_+_37992753 | 0.08 |
ENSDART00000193890
|
CR759843.2
|
|
| chr16_+_14707960 | 0.08 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr17_+_27545183 | 0.08 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr7_-_32875552 | 0.08 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr10_+_21807497 | 0.08 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr24_-_33308045 | 0.08 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr11_-_34522249 | 0.08 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr6_+_4872883 | 0.08 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr11_+_34522554 | 0.08 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr25_-_5119162 | 0.08 |
ENSDART00000153961
|
shisal1b
|
shisa like 1b |
| chr25_+_31323978 | 0.08 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr9_+_49727611 | 0.08 |
ENSDART00000110069
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr3_+_49521106 | 0.08 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr15_-_20731297 | 0.08 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr23_-_15090782 | 0.08 |
ENSDART00000133624
|
si:ch211-218g4.2
|
si:ch211-218g4.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.2 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.2 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.4 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.3 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0033335 | anal fin development(GO:0033335) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0048389 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.0 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.1 | GO:1903405 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.0 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.1 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.2 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |