Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
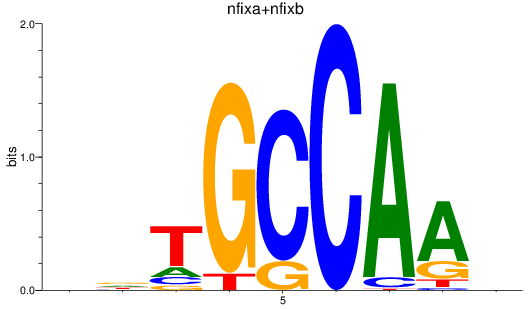
Results for nfixa+nfixb
Z-value: 1.11
Transcription factors associated with nfixa+nfixb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfixa
|
ENSDARG00000043226 | nuclear factor I/Xa |
|
nfixb
|
ENSDARG00000061836 | nuclear factor I/Xb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfixb | dr11_v1_chr3_+_14768364_14768364 | -0.86 | 5.9e-02 | Click! |
| nfixa | dr11_v1_chr1_+_51896147_51896147 | -0.43 | 4.7e-01 | Click! |
Activity profile of nfixa+nfixb motif
Sorted Z-values of nfixa+nfixb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_17672527 | 1.05 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr1_-_33645967 | 0.90 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr3_-_32541033 | 0.84 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_-_8030583 | 0.82 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr23_+_19790962 | 0.73 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr3_+_49021079 | 0.73 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr22_+_38229321 | 0.69 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr22_+_5478353 | 0.68 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr14_+_29975268 | 0.67 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr14_-_25444774 | 0.67 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr12_-_22524388 | 0.62 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr14_+_11458044 | 0.61 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr14_+_11457500 | 0.61 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr1_-_58900851 | 0.58 |
ENSDART00000183085
ENSDART00000188855 ENSDART00000182567 |
CABZ01084501.3
|
Danio rerio microfibril-associated glycoprotein 4-like (LOC100334800), transcript variant 2, mRNA. |
| chr19_+_14109348 | 0.58 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr11_-_3629201 | 0.54 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr2_-_14798295 | 0.53 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr22_-_15602760 | 0.53 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr22_+_25184459 | 0.53 |
ENSDART00000105308
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr3_-_39198113 | 0.52 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr14_-_21064199 | 0.52 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr2_-_3437862 | 0.51 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr13_-_9895564 | 0.50 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr7_-_8416750 | 0.48 |
ENSDART00000181857
|
jac1
|
jacalin 1 |
| chr22_+_19553390 | 0.48 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr16_-_22713152 | 0.47 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr1_-_54706039 | 0.46 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr9_+_38292947 | 0.46 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr15_-_31027112 | 0.45 |
ENSDART00000100185
|
lgals9l4
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 4 |
| chr19_+_10603405 | 0.44 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr22_-_15602593 | 0.44 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr16_+_7697878 | 0.43 |
ENSDART00000104176
ENSDART00000172977 |
ccr11.1
|
chemokine (C-C motif) receptor 11.1 |
| chr2_-_41861040 | 0.43 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr13_-_20381485 | 0.43 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr7_-_5207152 | 0.43 |
ENSDART00000172751
|
si:ch73-223f5.2
|
si:ch73-223f5.2 |
| chr3_-_24681404 | 0.41 |
ENSDART00000161612
|
BX569789.1
|
|
| chr3_+_32416948 | 0.40 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr1_-_9988840 | 0.40 |
ENSDART00000144565
|
si:dkeyp-75b4.9
|
si:dkeyp-75b4.9 |
| chr2_-_42375275 | 0.39 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr7_+_59649399 | 0.39 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr7_-_21928826 | 0.39 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr3_-_36750068 | 0.39 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr25_-_26018424 | 0.38 |
ENSDART00000089332
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_28082310 | 0.38 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr16_-_45209684 | 0.38 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr10_+_39263827 | 0.37 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr6_-_9952103 | 0.36 |
ENSDART00000065475
|
zp2l2
|
zona pellucida glycoprotein 2, like 2 |
| chr14_+_22076596 | 0.36 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr25_-_26018240 | 0.36 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr19_-_13962469 | 0.35 |
ENSDART00000159062
|
paqr7a
|
progestin and adipoQ receptor family member VII, a |
| chr16_+_38394371 | 0.34 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr1_-_999556 | 0.33 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr4_-_52189449 | 0.33 |
ENSDART00000192426
|
BX649431.4
|
|
| chr22_+_38767780 | 0.32 |
ENSDART00000149499
|
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr6_-_27108844 | 0.32 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr20_+_25581627 | 0.32 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr21_-_36619599 | 0.32 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr16_+_27444098 | 0.32 |
ENSDART00000157690
|
invs
|
inversin |
| chr19_-_12404590 | 0.32 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr6_+_40437987 | 0.31 |
ENSDART00000136487
|
ghrl
|
ghrelin/obestatin prepropeptide |
| chr14_+_15484544 | 0.31 |
ENSDART00000188649
|
CR382326.1
|
|
| chr20_+_26880668 | 0.31 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr6_-_15096556 | 0.30 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr18_+_3037998 | 0.30 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr13_+_15682803 | 0.29 |
ENSDART00000188063
|
CR931980.1
|
|
| chr19_-_9786914 | 0.29 |
ENSDART00000181669
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr3_-_55139127 | 0.29 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr4_-_8040436 | 0.28 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr6_+_39905021 | 0.28 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr11_-_24016761 | 0.28 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr3_-_22228602 | 0.28 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr21_+_19858627 | 0.28 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr11_+_14333441 | 0.27 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr5_-_23878809 | 0.27 |
ENSDART00000160603
|
si:ch211-135f11.4
|
si:ch211-135f11.4 |
| chr6_+_8630355 | 0.27 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr2_-_51757328 | 0.27 |
ENSDART00000189286
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr1_+_1789357 | 0.27 |
ENSDART00000006449
|
atp1a1a.2
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 2 |
| chr11_-_11575070 | 0.27 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr15_+_37986069 | 0.26 |
ENSDART00000156984
|
si:dkey-238d18.8
|
si:dkey-238d18.8 |
| chr3_-_26921475 | 0.26 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr25_+_16194450 | 0.26 |
ENSDART00000141994
|
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr3_-_40976288 | 0.25 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr19_-_7225060 | 0.25 |
ENSDART00000133179
ENSDART00000151138 ENSDART00000104799 ENSDART00000128331 |
col11a2
|
collagen, type XI, alpha 2 |
| chr5_-_37103487 | 0.25 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr25_-_30394746 | 0.25 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr11_-_11890001 | 0.25 |
ENSDART00000081544
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr2_-_14793343 | 0.25 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr11_+_43740949 | 0.24 |
ENSDART00000189296
|
CU862021.1
|
|
| chr19_-_42556086 | 0.24 |
ENSDART00000051731
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr25_-_16581233 | 0.24 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr13_+_18545819 | 0.24 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr9_+_15890558 | 0.24 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr15_+_38308421 | 0.24 |
ENSDART00000129941
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr2_-_36918709 | 0.23 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr7_+_25049545 | 0.23 |
ENSDART00000173896
ENSDART00000173566 |
si:dkey-23i12.7
ppp2r5b
|
si:dkey-23i12.7 protein phosphatase 2, regulatory subunit B', beta |
| chr7_+_12835048 | 0.23 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr13_+_18321140 | 0.23 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr3_-_8130926 | 0.22 |
ENSDART00000189495
|
si:ch211-51i16.1
|
si:ch211-51i16.1 |
| chr2_-_2642476 | 0.22 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr21_-_25555355 | 0.22 |
ENSDART00000144228
|
si:dkey-17e16.9
|
si:dkey-17e16.9 |
| chr6_+_32834760 | 0.22 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr1_-_9644630 | 0.22 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr16_+_48682309 | 0.22 |
ENSDART00000183957
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_+_48858132 | 0.22 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr5_-_10239079 | 0.22 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr14_+_40874608 | 0.22 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr4_+_37406676 | 0.22 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr7_+_61184551 | 0.22 |
ENSDART00000190788
|
zgc:194930
|
zgc:194930 |
| chr4_+_28997595 | 0.21 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr15_+_14154174 | 0.21 |
ENSDART00000161234
|
clca5.1
|
chloride channel accessory 5, tandem duplicate 1 |
| chr2_+_15069011 | 0.21 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr25_-_30429607 | 0.20 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr16_-_16619854 | 0.20 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr9_+_12890161 | 0.20 |
ENSDART00000146477
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr7_+_35238234 | 0.20 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr6_-_49078263 | 0.20 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr6_-_24301324 | 0.20 |
ENSDART00000171401
|
BX927081.1
|
|
| chr24_-_23998897 | 0.20 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr3_+_34988670 | 0.19 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr4_+_70556298 | 0.19 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr19_-_7144548 | 0.19 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr7_-_34294613 | 0.19 |
ENSDART00000191632
|
CR388195.1
|
|
| chr8_-_43834442 | 0.19 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr20_+_31985461 | 0.19 |
ENSDART00000191961
|
BX649312.1
|
|
| chr21_-_26558268 | 0.19 |
ENSDART00000065390
|
CR631122.1
|
|
| chr17_+_20589553 | 0.19 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr18_-_21177674 | 0.19 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr2_+_16191852 | 0.19 |
ENSDART00000152046
|
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr19_-_29832876 | 0.19 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr19_+_30990815 | 0.18 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr21_+_27448856 | 0.18 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr17_+_6382275 | 0.18 |
ENSDART00000056324
|
CR628323.2
|
|
| chr21_-_33126697 | 0.18 |
ENSDART00000189293
ENSDART00000084559 |
CU019662.1
|
|
| chr8_+_46536893 | 0.18 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr3_-_34068044 | 0.18 |
ENSDART00000187926
|
ighv10-1
|
immunoglobulin heavy variable 10-1 |
| chr5_-_26181863 | 0.17 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr23_+_16831063 | 0.17 |
ENSDART00000104782
|
zgc:173570
|
zgc:173570 |
| chr23_+_21414005 | 0.17 |
ENSDART00000144686
|
iffo2a
|
intermediate filament family orphan 2a |
| chr24_+_1294176 | 0.17 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr9_+_13999620 | 0.17 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr15_+_5280858 | 0.17 |
ENSDART00000174242
|
or121-1
|
odorant receptor, family E, subfamily 121, member 1 |
| chr19_-_24163845 | 0.17 |
ENSDART00000133277
|
prf1.2
|
perforin 1.2 |
| chr22_+_9918872 | 0.17 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr18_+_618005 | 0.17 |
ENSDART00000189667
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr9_-_30190513 | 0.17 |
ENSDART00000137083
|
impg2a
|
interphotoreceptor matrix proteoglycan 2a |
| chr7_+_61184104 | 0.16 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr21_-_31207522 | 0.16 |
ENSDART00000191637
|
zgc:152891
|
zgc:152891 |
| chr13_+_50375800 | 0.16 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr22_-_28653074 | 0.16 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr11_-_20956309 | 0.16 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr25_+_7494181 | 0.16 |
ENSDART00000165005
|
cat
|
catalase |
| chr17_-_8886735 | 0.16 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr19_-_5264517 | 0.16 |
ENSDART00000001821
|
prf1.3
|
perforin 1.3 |
| chr16_-_26727032 | 0.16 |
ENSDART00000177668
|
rnf41l
|
ring finger protein 41, like |
| chr21_+_15866522 | 0.15 |
ENSDART00000110728
|
gcnt4b
|
glucosaminyl (N-acetyl) transferase 4, core 2, b |
| chr8_+_20880848 | 0.15 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr5_-_38179220 | 0.15 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr21_+_25335807 | 0.15 |
ENSDART00000146443
|
or132-2
|
odorant receptor, family H, subfamily 132, member 2 |
| chr10_-_17988779 | 0.15 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr17_+_37249736 | 0.15 |
ENSDART00000189686
|
CR318625.1
|
|
| chr25_+_10953351 | 0.14 |
ENSDART00000154050
|
mhc1lda
|
major histocompatibility complex class I LDA |
| chr6_-_46782176 | 0.14 |
ENSDART00000185559
|
igfn1.4
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 4 |
| chr24_+_26432541 | 0.14 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr7_+_73801377 | 0.14 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr17_-_33707589 | 0.14 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr5_+_45918680 | 0.14 |
ENSDART00000036242
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr17_+_16873417 | 0.14 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr4_-_72100774 | 0.14 |
ENSDART00000170099
|
slco1f1
|
solute carrier organic anion transporter family, member 1F1 |
| chr15_+_5321612 | 0.14 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr24_+_15655233 | 0.14 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr8_-_31416403 | 0.13 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr5_+_31965845 | 0.13 |
ENSDART00000112968
|
myo1hb
|
myosin IHb |
| chr4_+_51347000 | 0.13 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr3_-_8765165 | 0.13 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr23_-_20361971 | 0.13 |
ENSDART00000133373
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr23_+_28381260 | 0.13 |
ENSDART00000162722
|
zgc:153867
|
zgc:153867 |
| chr24_-_20369760 | 0.13 |
ENSDART00000185585
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr2_-_24488652 | 0.13 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr9_-_39968820 | 0.13 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr7_+_21917857 | 0.12 |
ENSDART00000088041
|
ufsp1
|
UFM1-specific peptidase 1 (non-functional) |
| chr2_+_44518636 | 0.12 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr24_+_21174851 | 0.12 |
ENSDART00000154940
ENSDART00000155977 ENSDART00000122762 |
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr8_-_20138054 | 0.12 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr23_-_26880623 | 0.12 |
ENSDART00000038491
|
adcy6b
|
adenylate cyclase 6b |
| chr11_+_44579865 | 0.12 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr13_-_8815530 | 0.12 |
ENSDART00000122498
|
si:ch73-105b23.6
|
si:ch73-105b23.6 |
| chr10_+_36345176 | 0.12 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr22_+_10163901 | 0.12 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr16_-_17525322 | 0.12 |
ENSDART00000189720
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr14_-_9277152 | 0.12 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr19_-_24443867 | 0.12 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr2_-_51330414 | 0.12 |
ENSDART00000186450
|
si:dkeyp-104b3.21
|
si:dkeyp-104b3.21 |
| chr13_-_9024004 | 0.11 |
ENSDART00000169564
|
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr17_-_48743076 | 0.11 |
ENSDART00000173117
|
kcnk17
|
potassium channel, subfamily K, member 17 |
| chr5_+_26121393 | 0.11 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr7_-_59054322 | 0.11 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr15_+_31344472 | 0.11 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr12_-_6880694 | 0.11 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr11_-_1933793 | 0.10 |
ENSDART00000186850
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr2_-_37477654 | 0.10 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfixa+nfixb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.3 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.3 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 0.5 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.4 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.4 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.3 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.2 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 1.0 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0071962 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.3 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.0 | 0.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.2 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.0 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.5 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |