Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
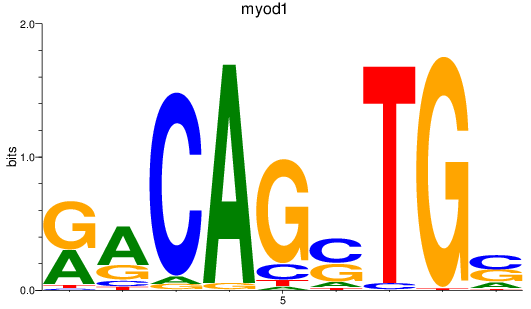
Results for myod1
Z-value: 2.32
Transcription factors associated with myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myod1
|
ENSDARG00000030110 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myod1 | dr11_v1_chr25_-_31423493_31423493 | 0.84 | 7.7e-02 | Click! |
Activity profile of myod1 motif
Sorted Z-values of myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_1051438 | 2.02 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr22_-_14128716 | 1.81 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr3_-_32817274 | 1.63 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr3_+_32526799 | 1.54 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_32526263 | 1.40 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr22_-_651719 | 1.37 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr2_+_55982940 | 1.37 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr6_-_42003780 | 1.30 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr20_-_27325258 | 1.24 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr9_-_98982 | 1.22 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr2_+_55982300 | 1.18 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr18_+_402048 | 1.14 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr6_-_39764995 | 1.12 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_31891110 | 1.12 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr1_-_38816685 | 1.12 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr12_-_26415499 | 1.09 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr12_-_4070058 | 1.09 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr9_-_22831836 | 1.09 |
ENSDART00000142585
|
neb
|
nebulin |
| chr16_-_31469065 | 1.08 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr12_-_28570989 | 1.08 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr16_-_12723738 | 1.06 |
ENSDART00000080414
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr21_+_27382893 | 1.04 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr2_-_44255537 | 1.02 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr9_-_7539297 | 0.99 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr6_-_39765546 | 0.97 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr2_-_42128714 | 0.97 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr2_-_21335131 | 0.96 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr17_+_27434626 | 0.96 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr13_+_23157053 | 0.94 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr9_-_48281941 | 0.94 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr13_-_7031033 | 0.94 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr14_+_31651533 | 0.94 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr2_-_31800521 | 0.91 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr24_-_33703504 | 0.89 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr22_-_10110959 | 0.88 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr7_-_69857692 | 0.88 |
ENSDART00000124764
|
myoz2a
|
myozenin 2a |
| chr14_-_17563773 | 0.86 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr16_+_25245857 | 0.84 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr13_+_22264914 | 0.83 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr18_+_29145681 | 0.82 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr22_-_26595027 | 0.81 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr23_-_5683147 | 0.81 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr23_+_43950674 | 0.80 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr6_-_35401282 | 0.79 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr22_+_16308450 | 0.78 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr19_+_46113828 | 0.77 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr8_-_18582922 | 0.77 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr14_+_31657412 | 0.76 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr23_-_32194397 | 0.75 |
ENSDART00000184206
ENSDART00000166682 |
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr18_-_6634424 | 0.74 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr16_-_12723324 | 0.74 |
ENSDART00000131915
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr22_+_16308806 | 0.73 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr14_-_29905962 | 0.72 |
ENSDART00000142605
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr16_-_14074594 | 0.72 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr8_-_11229523 | 0.71 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr25_+_14087045 | 0.71 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr11_-_45171139 | 0.71 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr20_+_26966725 | 0.71 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr7_+_31879649 | 0.70 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr16_+_12611362 | 0.70 |
ENSDART00000055160
|
il11a
|
interleukin 11a |
| chr12_+_18524953 | 0.70 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr23_-_31512496 | 0.70 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr7_+_31838320 | 0.69 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr13_+_42124566 | 0.69 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr8_+_44926946 | 0.68 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr6_+_3680651 | 0.68 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr24_-_41679356 | 0.67 |
ENSDART00000046821
|
lrrc30a
|
leucine rich repeat containing 30a |
| chr7_+_31879986 | 0.67 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_49397115 | 0.66 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr8_-_18200003 | 0.66 |
ENSDART00000080014
|
rps8b
|
ribosomal protein S8b |
| chr4_-_20181964 | 0.66 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr7_-_40578733 | 0.65 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr4_+_8670662 | 0.64 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr16_-_29146624 | 0.64 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr21_-_35419486 | 0.63 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr21_+_10866421 | 0.62 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr2_-_32457919 | 0.61 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr20_-_26866111 | 0.60 |
ENSDART00000077767
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr6_+_49028874 | 0.60 |
ENSDART00000175254
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr23_+_6077503 | 0.59 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr4_-_15603511 | 0.59 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr16_-_42894628 | 0.58 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr7_-_29292206 | 0.58 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr6_-_21189295 | 0.58 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr3_+_22242269 | 0.58 |
ENSDART00000168970
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr25_-_8602437 | 0.58 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr23_-_29003864 | 0.56 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr5_-_64355227 | 0.55 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr12_-_25380028 | 0.54 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr17_+_53311618 | 0.53 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr23_+_44614056 | 0.53 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr4_-_16412084 | 0.53 |
ENSDART00000188460
|
dcn
|
decorin |
| chr20_+_26967154 | 0.52 |
ENSDART00000153294
ENSDART00000132434 ENSDART00000143047 |
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr20_-_38758797 | 0.51 |
ENSDART00000061394
|
trim54
|
tripartite motif containing 54 |
| chr4_+_7841627 | 0.51 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr7_-_41858513 | 0.50 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr21_+_28749720 | 0.50 |
ENSDART00000145178
|
zgc:100829
|
zgc:100829 |
| chr14_+_6159356 | 0.50 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr4_-_4834347 | 0.50 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr7_+_65586624 | 0.49 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr14_-_33044955 | 0.49 |
ENSDART00000170626
|
kdrl
|
kinase insert domain receptor like |
| chr19_+_24488403 | 0.49 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr4_-_5597167 | 0.49 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr12_+_16967715 | 0.47 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr3_-_32169754 | 0.46 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr2_-_24289641 | 0.46 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr6_-_39051319 | 0.46 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr5_-_31904562 | 0.46 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr22_-_10541372 | 0.45 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr10_-_8295294 | 0.44 |
ENSDART00000075412
ENSDART00000163803 |
plpp1a
|
phospholipid phosphatase 1a |
| chr21_-_7265219 | 0.43 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr18_+_23249519 | 0.43 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr14_-_29906209 | 0.43 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr8_-_17980317 | 0.43 |
ENSDART00000129148
|
tnni3k
|
TNNI3 interacting kinase |
| chr20_-_21672970 | 0.42 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr22_-_10541712 | 0.42 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr13_-_24794486 | 0.42 |
ENSDART00000136177
|
slka
|
STE20-like kinase a |
| chr12_+_28749189 | 0.41 |
ENSDART00000013980
|
tbx21
|
T-box 21 |
| chr8_+_45294767 | 0.41 |
ENSDART00000191527
|
ubap2b
|
ubiquitin associated protein 2b |
| chr4_-_77624155 | 0.41 |
ENSDART00000099761
|
si:ch211-250m6.7
|
si:ch211-250m6.7 |
| chr3_-_31078348 | 0.40 |
ENSDART00000055360
|
elob
|
elongin B |
| chr14_-_25956804 | 0.40 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr23_-_9925568 | 0.40 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr5_+_37785152 | 0.40 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr11_+_24889170 | 0.40 |
ENSDART00000113310
|
lmod1b
|
leiomodin 1b (smooth muscle) |
| chr21_+_29077509 | 0.40 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr2_-_23768818 | 0.40 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr1_-_59141715 | 0.39 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr19_-_7450796 | 0.38 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr10_-_13178853 | 0.38 |
ENSDART00000163740
ENSDART00000166327 ENSDART00000160265 ENSDART00000164299 |
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr21_-_4250682 | 0.38 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr4_-_16354292 | 0.38 |
ENSDART00000139919
|
lum
|
lumican |
| chr14_+_22113331 | 0.38 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr14_-_30704075 | 0.37 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr3_-_31783737 | 0.37 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr7_+_20467549 | 0.37 |
ENSDART00000173724
|
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr5_-_30475011 | 0.37 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr2_-_6182098 | 0.37 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr7_-_5029478 | 0.36 |
ENSDART00000193819
|
ltb4r
|
leukotriene B4 receptor |
| chr1_+_50293938 | 0.36 |
ENSDART00000084184
|
aimp1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr1_-_47431453 | 0.36 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr9_+_30633184 | 0.35 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr6_-_10912424 | 0.34 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr11_+_27364338 | 0.34 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr11_-_5865744 | 0.34 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr10_-_8294965 | 0.34 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr18_+_25003207 | 0.34 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr8_+_47804284 | 0.34 |
ENSDART00000053285
|
ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 |
| chr17_+_38573471 | 0.34 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr16_+_3004422 | 0.34 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr7_+_13756374 | 0.33 |
ENSDART00000180808
|
rasl12
|
RAS-like, family 12 |
| chr9_-_49531762 | 0.33 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr17_+_15041647 | 0.33 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr11_+_11120532 | 0.33 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr15_+_28318005 | 0.33 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr25_+_16214854 | 0.33 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr10_+_26871751 | 0.33 |
ENSDART00000089205
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr11_+_6116096 | 0.33 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_-_1349922 | 0.32 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr5_+_6796291 | 0.32 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr18_+_23218980 | 0.32 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr5_-_51819027 | 0.32 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr6_-_7052408 | 0.32 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr22_+_18952781 | 0.32 |
ENSDART00000136390
|
hcn2b
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2b |
| chr6_+_43234213 | 0.32 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr3_-_31784082 | 0.31 |
ENSDART00000134201
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr9_-_23944470 | 0.31 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr24_+_20575259 | 0.31 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr17_-_45370200 | 0.31 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr17_+_53311243 | 0.31 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr17_-_34963575 | 0.31 |
ENSDART00000145664
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr16_-_33816082 | 0.31 |
ENSDART00000181769
|
rspo1
|
R-spondin 1 |
| chr21_-_43126881 | 0.31 |
ENSDART00000174463
ENSDART00000160845 |
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr2_+_6181383 | 0.30 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr6_-_48082525 | 0.30 |
ENSDART00000192049
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr1_-_14233815 | 0.30 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr8_+_23194212 | 0.30 |
ENSDART00000028946
ENSDART00000186842 ENSDART00000137815 ENSDART00000136914 ENSDART00000144855 ENSDART00000182689 |
tpd52l2a
|
tumor protein D52-like 2a |
| chr3_+_1096831 | 0.30 |
ENSDART00000132817
|
si:ch1073-322p19.1
|
si:ch1073-322p19.1 |
| chr22_+_30335936 | 0.30 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr12_+_23866368 | 0.29 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr22_+_11756040 | 0.29 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr1_-_10841348 | 0.29 |
ENSDART00000148305
|
dmd
|
dystrophin |
| chr16_+_46401576 | 0.29 |
ENSDART00000130264
|
rpz
|
rapunzel |
| chr6_+_40629066 | 0.29 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr2_-_25120197 | 0.29 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr7_+_73690366 | 0.29 |
ENSDART00000191494
ENSDART00000123429 ENSDART00000173403 |
si:dkey-46i9.6
|
si:dkey-46i9.6 |
| chr19_-_25519612 | 0.29 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr7_+_55112922 | 0.29 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr21_-_25741096 | 0.29 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr13_+_1575276 | 0.29 |
ENSDART00000165987
|
DST
|
dystonin |
| chr5_+_37854685 | 0.28 |
ENSDART00000051222
ENSDART00000185283 |
ins
|
preproinsulin |
| chr11_-_43226255 | 0.28 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_+_24468922 | 0.28 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_-_50287949 | 0.28 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr21_+_40498628 | 0.28 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr15_-_12500938 | 0.28 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr7_-_22632518 | 0.27 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr10_+_6496185 | 0.27 |
ENSDART00000164770
|
reep5
|
receptor accessory protein 5 |
| chr24_+_26276805 | 0.27 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr5_-_48307804 | 0.27 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
Network of associatons between targets according to the STRING database.
First level regulatory network of myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.6 | 3.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.5 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.3 | 1.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.3 | 2.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.2 | 1.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.2 | 2.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 2.1 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.7 | GO:0060307 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.2 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.6 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.6 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.8 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.9 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.4 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.3 | GO:2000252 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.7 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 1.1 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.5 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.2 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 1.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.9 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.1 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 1.0 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.5 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.4 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.4 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.0 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0033152 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.0 | GO:0003230 | cardiac atrium development(GO:0003230) |
| 0.0 | 0.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 5.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.4 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 7.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 2.9 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.0 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.3 | 1.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 3.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 0.8 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.3 | 0.8 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.1 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.7 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 0.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.8 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.9 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.0 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |