Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
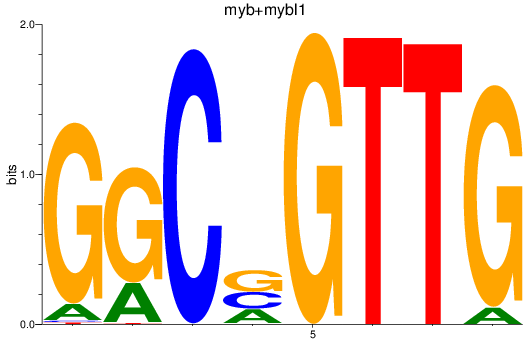
Results for myb+mybl1
Z-value: 1.81
Transcription factors associated with myb+mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mybl1
|
ENSDARG00000030999 | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
|
myb
|
ENSDARG00000053666 | v-myb avian myeloblastosis viral oncogene homolog |
|
myb
|
ENSDARG00000114024 | v-myb avian myeloblastosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl1 | dr11_v1_chr24_+_23791758_23791839 | 0.83 | 7.9e-02 | Click! |
| myb | dr11_v1_chr23_+_31815423_31815492 | -0.62 | 2.6e-01 | Click! |
Activity profile of myb+mybl1 motif
Sorted Z-values of myb+mybl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_101758 | 2.01 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr3_+_19609638 | 1.39 |
ENSDART00000079319
|
rprml
|
reprimo-like |
| chr3_-_12890670 | 1.34 |
ENSDART00000159934
ENSDART00000188607 |
btbd17b
|
BTB (POZ) domain containing 17b |
| chr7_+_29148714 | 1.23 |
ENSDART00000170342
|
mtbl
|
metallothionein-B-like |
| chr12_+_9880493 | 1.19 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr10_-_7974155 | 1.14 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr24_-_25574967 | 1.11 |
ENSDART00000189828
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr19_-_3240605 | 1.06 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr16_-_29458806 | 1.05 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr9_-_41277347 | 0.98 |
ENSDART00000181213
|
glsb
|
glutaminase b |
| chr13_-_27660955 | 0.94 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr11_-_36957127 | 0.91 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr10_+_21576909 | 0.90 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr1_+_36436936 | 0.89 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr1_+_12763920 | 0.89 |
ENSDART00000189465
|
pcdh10a
|
protocadherin 10a |
| chr10_+_2234283 | 0.88 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr24_-_7699356 | 0.87 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr5_-_24000211 | 0.85 |
ENSDART00000188865
|
map7d2a
|
MAP7 domain containing 2a |
| chr22_+_12790837 | 0.84 |
ENSDART00000063055
|
mstna
|
myostatin a |
| chr10_+_34685135 | 0.84 |
ENSDART00000184999
|
nbeaa
|
neurobeachin a |
| chr21_-_12036134 | 0.83 |
ENSDART00000031658
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr11_-_13341483 | 0.83 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr10_+_11038192 | 0.80 |
ENSDART00000047954
|
ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr10_+_10636237 | 0.80 |
ENSDART00000136853
|
fam163b
|
family with sequence similarity 163, member B |
| chr24_-_4973765 | 0.79 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr19_-_38872650 | 0.78 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr11_-_13341051 | 0.78 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr17_-_20711735 | 0.77 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr3_+_46459540 | 0.77 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr13_+_46718518 | 0.77 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr1_+_14073891 | 0.77 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr12_+_42436920 | 0.76 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr17_-_37156520 | 0.76 |
ENSDART00000145669
|
dtnbb
|
dystrobrevin, beta b |
| chr9_+_7456076 | 0.75 |
ENSDART00000125824
ENSDART00000122526 |
tmem198a
|
transmembrane protein 198a |
| chr1_-_38813679 | 0.73 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr5_-_45773776 | 0.73 |
ENSDART00000124336
|
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr5_-_23999777 | 0.71 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr24_-_25673405 | 0.71 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr19_+_37701450 | 0.71 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr25_-_20381271 | 0.69 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr10_-_25860102 | 0.69 |
ENSDART00000080789
|
trpc4a
|
transient receptor potential cation channel, subfamily C, member 4a |
| chr22_+_413349 | 0.68 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr20_-_20930926 | 0.67 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr1_-_44701313 | 0.66 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr23_+_23187442 | 0.65 |
ENSDART00000184920
|
klhl17
|
kelch-like family member 17 |
| chr20_+_34596954 | 0.65 |
ENSDART00000076946
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr7_-_49646251 | 0.65 |
ENSDART00000193674
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr18_-_47533692 | 0.65 |
ENSDART00000191097
|
CABZ01112461.1
|
|
| chr19_+_8144556 | 0.65 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr10_+_21737745 | 0.64 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr17_-_45552602 | 0.64 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr16_-_23379464 | 0.64 |
ENSDART00000045891
|
trim46a
|
tripartite motif containing 46a |
| chr2_+_38554260 | 0.64 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr5_-_26466169 | 0.63 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr20_-_45423498 | 0.63 |
ENSDART00000098424
|
trib2
|
tribbles pseudokinase 2 |
| chr16_+_22618620 | 0.63 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr14_-_33177935 | 0.63 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr5_-_46980651 | 0.62 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr5_+_44064764 | 0.62 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr11_+_38280454 | 0.62 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr17_-_2386569 | 0.61 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr2_+_46032678 | 0.61 |
ENSDART00000184382
ENSDART00000125971 |
gpc1b
|
glypican 1b |
| chr15_+_29393519 | 0.61 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr5_-_32424402 | 0.61 |
ENSDART00000077549
|
ncs1a
|
neuronal calcium sensor 1a |
| chr7_+_71547981 | 0.61 |
ENSDART00000012070
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr1_-_40608392 | 0.61 |
ENSDART00000101671
|
clgn
|
calmegin |
| chr1_+_27808916 | 0.60 |
ENSDART00000102335
|
pspc1
|
paraspeckle component 1 |
| chr8_-_24135171 | 0.59 |
ENSDART00000112926
|
adora1b
|
adenosine A1 receptor b |
| chr12_+_30046320 | 0.58 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr3_-_36115339 | 0.57 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr3_+_51563695 | 0.57 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr11_-_6188413 | 0.56 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr3_-_46811611 | 0.56 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr23_-_36724575 | 0.56 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_+_45456066 | 0.56 |
ENSDART00000093365
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr15_-_13254480 | 0.55 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr19_+_21266008 | 0.54 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr2_+_1002104 | 0.54 |
ENSDART00000159162
|
cacna1eb
|
calcium channel, voltage-dependent, R type, alpha 1E subunit b |
| chr5_+_20257225 | 0.54 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr11_-_472547 | 0.53 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr2_+_35240485 | 0.53 |
ENSDART00000179804
|
tnr
|
tenascin R (restrictin, janusin) |
| chr22_+_28803739 | 0.53 |
ENSDART00000129476
ENSDART00000189726 |
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr10_+_7564106 | 0.53 |
ENSDART00000159042
|
purg
|
purine-rich element binding protein G |
| chr3_-_59981476 | 0.52 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr22_+_34430310 | 0.52 |
ENSDART00000109860
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr25_-_518656 | 0.51 |
ENSDART00000156421
|
myo9ab
|
myosin IXAb |
| chr19_+_41701660 | 0.51 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr8_+_29749017 | 0.51 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr6_-_40744720 | 0.51 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr25_+_13791627 | 0.51 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr2_-_11662851 | 0.51 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr24_-_35707552 | 0.51 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_-_31915423 | 0.51 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr17_+_10593398 | 0.50 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr6_-_36552844 | 0.50 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr1_-_47089818 | 0.50 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr20_-_53981626 | 0.50 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr23_-_7125494 | 0.49 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr12_+_40810418 | 0.49 |
ENSDART00000183393
|
CDH18
|
cadherin 18 |
| chr6_+_18569453 | 0.49 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr9_-_30808073 | 0.49 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr5_+_59392183 | 0.49 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr2_-_16224083 | 0.48 |
ENSDART00000165953
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_20756013 | 0.48 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr9_+_44397151 | 0.48 |
ENSDART00000185947
|
ssfa2
|
sperm specific antigen 2 |
| chr7_+_63324888 | 0.47 |
ENSDART00000147785
|
pcdh7b
|
protocadherin 7b |
| chr5_+_20147830 | 0.47 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr14_-_48588422 | 0.47 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr6_-_18128721 | 0.46 |
ENSDART00000160036
|
SEC14L1
|
si:dkey-237i9.1 |
| chr13_+_3819475 | 0.46 |
ENSDART00000139958
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr22_+_24157807 | 0.46 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr19_-_28789404 | 0.46 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr24_+_32472155 | 0.46 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr23_-_32390742 | 0.45 |
ENSDART00000032861
|
ankrd52a
|
ankyrin repeat domain 52a |
| chr4_-_7811925 | 0.45 |
ENSDART00000103070
|
cdk17
|
cyclin-dependent kinase 17 |
| chr23_+_28092083 | 0.45 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr21_-_17482465 | 0.45 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr7_+_20109968 | 0.45 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr1_+_604127 | 0.45 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr22_-_25033105 | 0.44 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr19_+_38167468 | 0.44 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr8_+_49149907 | 0.44 |
ENSDART00000138746
|
snpha
|
syntaphilin a |
| chr17_+_18117976 | 0.44 |
ENSDART00000123745
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr25_-_16666886 | 0.44 |
ENSDART00000155764
|
slc25a18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr6_-_30658755 | 0.43 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr24_-_37472727 | 0.43 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr11_+_14622379 | 0.43 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr25_-_554142 | 0.43 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr14_-_33081784 | 0.43 |
ENSDART00000165726
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr15_+_36977208 | 0.43 |
ENSDART00000183625
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr24_-_28437833 | 0.43 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr20_-_3390406 | 0.43 |
ENSDART00000136987
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_+_37750016 | 0.43 |
ENSDART00000154726
|
si:dkeyp-66d1.7
|
si:dkeyp-66d1.7 |
| chr12_+_17436904 | 0.42 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr5_+_6892195 | 0.42 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr23_+_22335407 | 0.42 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr1_-_45920632 | 0.42 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr17_+_18117358 | 0.42 |
ENSDART00000144894
|
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr14_-_33083539 | 0.42 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr3_+_39853788 | 0.42 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr13_-_33009734 | 0.42 |
ENSDART00000134140
|
rbm25a
|
RNA binding motif protein 25a |
| chr21_+_33249478 | 0.42 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr3_+_17456428 | 0.41 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr1_-_44704261 | 0.41 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr15_+_15786160 | 0.41 |
ENSDART00000130670
ENSDART00000090939 |
tada2a
|
transcriptional adaptor 2A |
| chr17_+_23938283 | 0.40 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr15_-_45246911 | 0.40 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr18_+_7639401 | 0.39 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr7_+_20110336 | 0.39 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr20_-_18731268 | 0.39 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr10_+_20128267 | 0.38 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr11_+_41560792 | 0.38 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr16_+_5612547 | 0.38 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr7_+_71547747 | 0.38 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr13_-_23756700 | 0.38 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr18_+_15132112 | 0.38 |
ENSDART00000099764
|
zgc:153031
|
zgc:153031 |
| chr4_-_8611841 | 0.37 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr2_+_46411079 | 0.37 |
ENSDART00000187016
|
FQ377603.1
|
|
| chr3_+_24459709 | 0.37 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr23_-_21814739 | 0.37 |
ENSDART00000190409
|
ephb2a
|
eph receptor B2a |
| chr24_-_33780387 | 0.37 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr17_+_21818093 | 0.36 |
ENSDART00000125335
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr1_+_49878000 | 0.36 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr15_+_16525126 | 0.36 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr19_-_41991104 | 0.36 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr16_-_29164379 | 0.35 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr10_-_28368695 | 0.35 |
ENSDART00000179416
|
cltca
|
clathrin, heavy chain a (Hc) |
| chr3_-_59981162 | 0.35 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr16_+_32238020 | 0.35 |
ENSDART00000017562
|
klhl32
|
kelch-like family member 32 |
| chr21_-_30273418 | 0.34 |
ENSDART00000187069
ENSDART00000181492 |
zgc:175066
|
zgc:175066 |
| chr16_+_53125918 | 0.34 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr17_+_12865746 | 0.34 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr25_-_20666328 | 0.34 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr1_-_55196103 | 0.33 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr7_-_72427616 | 0.33 |
ENSDART00000181878
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr1_-_46718353 | 0.33 |
ENSDART00000074514
|
spryd7a
|
SPRY domain containing 7a |
| chr7_+_53754653 | 0.33 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr8_+_34922981 | 0.33 |
ENSDART00000188445
ENSDART00000188818 ENSDART00000182965 ENSDART00000180718 |
BX005361.1
|
|
| chr22_-_20342260 | 0.32 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr1_+_19434198 | 0.32 |
ENSDART00000012552
|
clockb
|
clock circadian regulator b |
| chr16_+_41717313 | 0.32 |
ENSDART00000015170
ENSDART00000075937 |
mtdhb
|
metadherin b |
| chr3_-_26524934 | 0.32 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr24_-_12749116 | 0.32 |
ENSDART00000093583
|
cox19
|
COX19 cytochrome c oxidase assembly factor |
| chr13_+_21870269 | 0.32 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr6_+_58280936 | 0.31 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr9_-_24209083 | 0.31 |
ENSDART00000134599
|
zgc:153521
|
zgc:153521 |
| chr13_+_40034176 | 0.31 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr2_+_30969029 | 0.31 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr7_+_56253914 | 0.30 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr2_-_42827336 | 0.30 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr18_+_45889496 | 0.30 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr5_+_21931124 | 0.30 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr21_-_4539899 | 0.30 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr21_-_26691959 | 0.29 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr10_-_28368913 | 0.29 |
ENSDART00000063849
|
cltca
|
clathrin, heavy chain a (Hc) |
| chr11_+_16152316 | 0.29 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr1_+_44540293 | 0.29 |
ENSDART00000003497
ENSDART00000179698 |
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr15_-_23942861 | 0.29 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr13_-_4303641 | 0.29 |
ENSDART00000136262
|
ppp2r5d
|
protein phosphatase 2, regulatory subunit B', delta |
| chr21_-_38717854 | 0.28 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of myb+mybl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.2 | 0.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.9 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 1.0 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 1.0 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.6 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.6 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.2 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.5 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.2 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 1.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.9 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.4 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 1.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 1.0 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of mast cell activation(GO:0033003) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.9 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.1 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.9 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.2 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 2.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |