Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
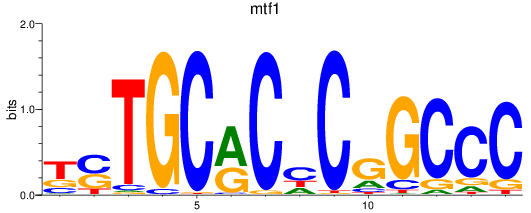
Results for mtf1
Z-value: 1.00
Transcription factors associated with mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mtf1
|
ENSDARG00000102898 | metal-regulatory transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mtf1 | dr11_v1_chr16_+_4133519_4133519 | 0.65 | 2.3e-01 | Click! |
Activity profile of mtf1 motif
Sorted Z-values of mtf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_59251874 | 0.94 |
ENSDART00000168919
|
olfm2b
|
olfactomedin 2b |
| chr3_-_50124413 | 0.94 |
ENSDART00000189920
|
cldnk
|
claudin k |
| chr24_-_38384432 | 0.81 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr16_-_29452039 | 0.75 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr22_+_5574952 | 0.69 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr10_+_21789954 | 0.68 |
ENSDART00000157769
ENSDART00000171703 |
pcdh1gc5
|
protocadherin 1 gamma c 5 |
| chr1_-_53750522 | 0.67 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr1_+_50085440 | 0.64 |
ENSDART00000018469
ENSDART00000134988 |
npnt
|
nephronectin |
| chr1_-_12278522 | 0.62 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr21_+_3093419 | 0.60 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr1_-_12278056 | 0.60 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr1_+_50085640 | 0.60 |
ENSDART00000158988
ENSDART00000165973 |
npnt
|
nephronectin |
| chr23_-_3681026 | 0.59 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr4_+_23223881 | 0.58 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr22_-_38607504 | 0.58 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr12_+_22745618 | 0.56 |
ENSDART00000181024
ENSDART00000183993 ENSDART00000077548 |
ablim2
|
actin binding LIM protein family, member 2 |
| chr3_-_56933578 | 0.55 |
ENSDART00000192185
|
hid1a
|
HID1 domain containing a |
| chr17_-_12385308 | 0.53 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_+_36614196 | 0.50 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr4_-_23643272 | 0.50 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr8_-_39977026 | 0.49 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr14_+_44588353 | 0.49 |
ENSDART00000173421
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr12_+_41510492 | 0.48 |
ENSDART00000170976
ENSDART00000176164 |
kif5bb
|
kinesin family member 5B, b |
| chr20_+_27298783 | 0.47 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr19_+_37701450 | 0.47 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr7_+_25323742 | 0.47 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr6_-_20875111 | 0.46 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr13_-_23756700 | 0.44 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr6_+_10450000 | 0.44 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr12_+_41991635 | 0.43 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr14_+_597532 | 0.42 |
ENSDART00000159805
|
LO018568.2
|
|
| chr23_-_28239750 | 0.42 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr24_-_28333029 | 0.41 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr17_+_51499789 | 0.41 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr17_-_36988455 | 0.40 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr19_-_19556778 | 0.39 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr23_+_45611649 | 0.38 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr22_-_37349967 | 0.38 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr2_+_33796207 | 0.38 |
ENSDART00000124647
|
kiss1ra
|
KISS1 receptor a |
| chr15_-_18429550 | 0.37 |
ENSDART00000136208
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr1_+_9004719 | 0.35 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr14_-_33177935 | 0.33 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr6_+_54498220 | 0.32 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr24_-_19718077 | 0.32 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr24_-_37877743 | 0.31 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr23_+_23485858 | 0.29 |
ENSDART00000114067
|
agrn
|
agrin |
| chr20_-_46362606 | 0.28 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr3_+_53052769 | 0.28 |
ENSDART00000156935
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr2_+_54641644 | 0.28 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr17_-_125091 | 0.28 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr17_+_48831579 | 0.28 |
ENSDART00000154824
ENSDART00000178697 ENSDART00000192703 ENSDART00000180378 |
daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr5_-_45651548 | 0.27 |
ENSDART00000097645
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr2_+_33457310 | 0.27 |
ENSDART00000056657
|
zgc:113531
|
zgc:113531 |
| chr6_+_7553085 | 0.27 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr13_+_52006253 | 0.26 |
ENSDART00000181259
|
CABZ01080135.1
|
|
| chr23_+_20931030 | 0.26 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr13_-_36184476 | 0.26 |
ENSDART00000057185
|
map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_+_49246099 | 0.26 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr14_-_45394704 | 0.26 |
ENSDART00000173078
ENSDART00000125970 |
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr7_-_38861741 | 0.25 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr19_+_43017931 | 0.25 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr8_-_51524446 | 0.24 |
ENSDART00000167394
ENSDART00000147742 |
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr14_+_5117072 | 0.24 |
ENSDART00000189628
|
nanos1
|
nanos homolog 1 |
| chr15_-_44275069 | 0.23 |
ENSDART00000124980
|
CU929391.2
|
|
| chr13_-_27621246 | 0.23 |
ENSDART00000102232
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr1_+_44906517 | 0.22 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr8_+_31435452 | 0.22 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr18_+_8231138 | 0.22 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr20_+_700616 | 0.22 |
ENSDART00000168166
|
senp6a
|
SUMO1/sentrin specific peptidase 6a |
| chr20_-_3403033 | 0.21 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr22_+_26703026 | 0.21 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr13_-_27620815 | 0.21 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr15_+_2559875 | 0.20 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr25_-_36020344 | 0.19 |
ENSDART00000181448
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr2_+_327081 | 0.18 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr2_-_51095743 | 0.17 |
ENSDART00000184646
|
THEM6
|
si:ch73-52e5.2 |
| chr8_-_53926228 | 0.17 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr23_+_39413163 | 0.17 |
ENSDART00000184254
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr23_-_18057553 | 0.16 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr2_-_16216568 | 0.16 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr16_-_42152145 | 0.16 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr22_-_20950448 | 0.16 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr11_-_3334248 | 0.16 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr24_-_37877554 | 0.16 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr10_+_5954787 | 0.15 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr24_+_28525507 | 0.15 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr1_-_35928942 | 0.14 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr4_+_8376362 | 0.14 |
ENSDART00000138653
ENSDART00000132647 |
erc1b
|
ELKS/RAB6-interacting/CAST family member 1b |
| chr9_-_48397702 | 0.14 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr12_+_20149707 | 0.13 |
ENSDART00000181942
|
foxj1b
|
forkhead box J1b |
| chr12_+_48851381 | 0.13 |
ENSDART00000187241
ENSDART00000187796 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr23_-_45682136 | 0.13 |
ENSDART00000164646
|
fam160a1b
|
family with sequence similarity 160, member A1b |
| chr8_+_26329482 | 0.13 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr15_+_3219134 | 0.13 |
ENSDART00000113532
|
LO017656.1
|
|
| chr18_-_7454422 | 0.12 |
ENSDART00000124709
|
pdp2
|
putative pyruvate dehydrogenase phosphatase isoenzyme 2 |
| chr24_+_10397865 | 0.12 |
ENSDART00000155557
|
si:ch211-69l10.4
|
si:ch211-69l10.4 |
| chr19_-_35400819 | 0.12 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr23_+_45611980 | 0.12 |
ENSDART00000181582
|
dclk2b
|
doublecortin-like kinase 2b |
| chr25_+_3008263 | 0.11 |
ENSDART00000160484
|
neo1b
|
neogenin 1b |
| chr6_+_31684 | 0.11 |
ENSDART00000188853
ENSDART00000184553 |
CZQB01141835.2
|
|
| chr9_-_128036 | 0.10 |
ENSDART00000165960
|
FQ377903.2
|
|
| chr18_+_18000887 | 0.10 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr5_-_14211487 | 0.09 |
ENSDART00000135731
|
npffr1l2
|
neuropeptide FF receptor 1 like 2 |
| chr24_-_3477103 | 0.09 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr6_+_11397269 | 0.09 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr11_-_575786 | 0.09 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr7_-_74090168 | 0.09 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr19_-_11336782 | 0.08 |
ENSDART00000131014
|
sept7a
|
septin 7a |
| chr8_+_8973425 | 0.08 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr7_-_50604626 | 0.07 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr20_-_48470599 | 0.07 |
ENSDART00000166857
|
CABZ01059120.1
|
|
| chr20_-_1314537 | 0.07 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr14_-_3070613 | 0.07 |
ENSDART00000193729
ENSDART00000090196 |
slc35a4
|
solute carrier family 35, member A4 |
| chr3_+_23221047 | 0.07 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr10_-_26997344 | 0.06 |
ENSDART00000146595
|
zgc:193742
|
zgc:193742 |
| chr4_+_53268458 | 0.06 |
ENSDART00000165335
|
si:dkey-250k10.1
|
si:dkey-250k10.1 |
| chr7_+_59169081 | 0.06 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr3_+_18756231 | 0.05 |
ENSDART00000089679
|
narfl
|
nuclear prelamin A recognition factor-like |
| chr20_+_560805 | 0.05 |
ENSDART00000152149
|
si:dkey-121j17.6
|
si:dkey-121j17.6 |
| chr2_+_29257942 | 0.04 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr1_-_44000940 | 0.04 |
ENSDART00000134932
ENSDART00000138447 |
zgc:66472
|
zgc:66472 |
| chr2_-_43739740 | 0.04 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr17_+_37215820 | 0.03 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr16_+_11029762 | 0.03 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr2_+_314249 | 0.03 |
ENSDART00000082086
|
zgc:113452
|
zgc:113452 |
| chr20_+_39250673 | 0.03 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr15_-_43284021 | 0.02 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr25_+_17313568 | 0.02 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr11_-_18017287 | 0.01 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr14_-_41075262 | 0.01 |
ENSDART00000180518
|
drp2
|
dystrophin related protein 2 |
| chr20_-_36408836 | 0.01 |
ENSDART00000076419
|
lbr
|
lamin B receptor |
| chr6_-_7686594 | 0.01 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr6_-_37513133 | 0.01 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| chr1_-_35929143 | 0.00 |
ENSDART00000185002
|
smad1
|
SMAD family member 1 |
| chr19_+_1005933 | 0.00 |
ENSDART00000191953
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of mtf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.5 | GO:0035992 | tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.7 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.3 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.4 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 0.3 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 | 1.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.5 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |