Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
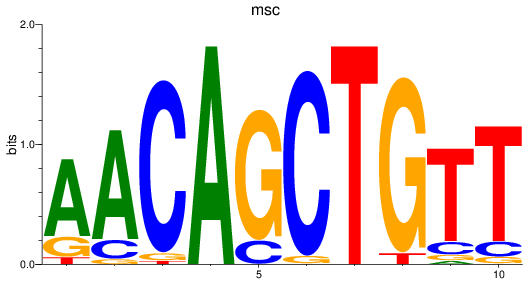
Results for msc
Z-value: 1.30
Transcription factors associated with msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msc
|
ENSDARG00000110016 | musculin (activated B-cell factor-1) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msc | dr11_v1_chr24_+_13735616_13735616 | 0.12 | 8.5e-01 | Click! |
Activity profile of msc motif
Sorted Z-values of msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43952958 | 0.88 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr6_+_13742899 | 0.83 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr13_+_24834199 | 0.78 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr20_-_27325258 | 0.70 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr13_+_42124566 | 0.69 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr21_+_27382893 | 0.65 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr20_+_35382482 | 0.65 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_-_38451082 | 0.64 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr14_-_32255126 | 0.64 |
ENSDART00000017259
|
fgf13a
|
fibroblast growth factor 13a |
| chr4_-_75175407 | 0.63 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr13_+_1575276 | 0.61 |
ENSDART00000165987
|
DST
|
dystonin |
| chr11_-_1291012 | 0.61 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr21_+_11468642 | 0.60 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr1_+_25801648 | 0.59 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr7_+_7048245 | 0.57 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr15_-_32383529 | 0.57 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr15_+_40188076 | 0.56 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr7_+_6969909 | 0.56 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr12_-_28570989 | 0.56 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr1_-_25911292 | 0.53 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr3_-_36115339 | 0.52 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr9_+_40939336 | 0.51 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr1_+_52560549 | 0.51 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr14_+_30730749 | 0.50 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr15_-_15357178 | 0.48 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr7_+_27059330 | 0.47 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr5_-_51819027 | 0.47 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr17_+_16873417 | 0.47 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr7_+_31879649 | 0.45 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr2_-_42128714 | 0.45 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr1_-_54997746 | 0.43 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr21_+_11468934 | 0.43 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr6_+_23887314 | 0.43 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr8_-_14151051 | 0.41 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr1_+_2101541 | 0.41 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr7_+_31879986 | 0.41 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr15_-_32383340 | 0.41 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr4_-_9586713 | 0.41 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr5_-_31904562 | 0.41 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr21_-_16632808 | 0.40 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr19_-_9711472 | 0.39 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr10_+_34315719 | 0.39 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr18_+_25003207 | 0.38 |
ENSDART00000099476
ENSDART00000132285 |
fam174b
|
family with sequence similarity 174, member B |
| chr19_+_1184878 | 0.37 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr14_+_21783229 | 0.37 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr14_+_7939398 | 0.37 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr1_+_33401774 | 0.37 |
ENSDART00000149786
ENSDART00000110026 ENSDART00000148946 |
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr19_-_10425140 | 0.37 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr23_-_18057553 | 0.36 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr7_-_23745984 | 0.36 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr2_+_45696743 | 0.36 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr10_+_15777258 | 0.35 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr21_-_20929575 | 0.35 |
ENSDART00000163889
|
c6
|
complement component 6 |
| chr14_+_5936996 | 0.35 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr9_+_52492639 | 0.35 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr17_-_14726824 | 0.35 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr2_-_44255537 | 0.34 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr13_-_31452516 | 0.34 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr1_-_20593778 | 0.34 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr14_+_7939216 | 0.34 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr10_-_34741738 | 0.34 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr8_-_44004135 | 0.34 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr7_+_26224211 | 0.33 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr5_+_3501859 | 0.33 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr13_-_12581388 | 0.33 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr14_+_21783400 | 0.33 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr5_+_42097608 | 0.32 |
ENSDART00000014404
|
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr16_+_3004422 | 0.31 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr10_+_15777064 | 0.31 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr5_-_22019061 | 0.31 |
ENSDART00000113066
|
amer1
|
APC membrane recruitment protein 1 |
| chr2_+_37480669 | 0.31 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr6_-_40098641 | 0.30 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr9_-_98982 | 0.30 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr20_+_23501535 | 0.29 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr7_+_19552381 | 0.29 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr5_-_30475011 | 0.28 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr21_-_27123139 | 0.28 |
ENSDART00000077478
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr24_+_38155830 | 0.28 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr18_+_39028417 | 0.28 |
ENSDART00000148428
|
myo5aa
|
myosin VAa |
| chr2_-_30659222 | 0.27 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr11_+_24314148 | 0.27 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_+_39222598 | 0.27 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr8_+_25247245 | 0.27 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr6_+_27339962 | 0.27 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr13_-_40499296 | 0.27 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr5_-_40734045 | 0.27 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr4_+_12615836 | 0.27 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr23_-_30954738 | 0.27 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr13_-_31164749 | 0.27 |
ENSDART00000049180
|
mapk8a
|
mitogen-activated protein kinase 8a |
| chr23_-_24263474 | 0.26 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr11_+_24313931 | 0.26 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr20_-_26467307 | 0.26 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr12_+_5209822 | 0.26 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr6_+_40587551 | 0.25 |
ENSDART00000155554
|
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr12_+_33038757 | 0.25 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr6_-_16667886 | 0.25 |
ENSDART00000180854
ENSDART00000190116 |
unc80
|
unc-80 homolog (C. elegans) |
| chr4_+_12617108 | 0.25 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr23_-_8373676 | 0.25 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr14_+_26805388 | 0.25 |
ENSDART00000044389
ENSDART00000173126 |
klhl4
|
kelch-like family member 4 |
| chr2_-_32551178 | 0.25 |
ENSDART00000145603
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr23_-_32162810 | 0.25 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr8_-_43997538 | 0.25 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr5_-_10768258 | 0.24 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr23_+_35714574 | 0.24 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr12_+_17504559 | 0.24 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr1_-_17650223 | 0.24 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr9_+_23665777 | 0.24 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr21_-_3770636 | 0.24 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr24_+_33802528 | 0.24 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr14_-_16754262 | 0.24 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr1_-_29747702 | 0.24 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr7_+_27277236 | 0.24 |
ENSDART00000185161
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_-_10432134 | 0.24 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr21_-_17482465 | 0.23 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr25_+_36674715 | 0.23 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr4_+_20255160 | 0.23 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr17_-_14836320 | 0.23 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr9_-_28939181 | 0.23 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr6_+_2097690 | 0.23 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr18_-_5781922 | 0.23 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr14_-_15699528 | 0.23 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr18_-_399554 | 0.23 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr3_+_29714775 | 0.22 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr21_-_24865217 | 0.22 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr7_+_63325819 | 0.22 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr14_-_46832825 | 0.22 |
ENSDART00000149571
|
ldb2a
|
LIM domain binding 2a |
| chr8_-_11229523 | 0.22 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr24_+_24461558 | 0.22 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr19_-_10214264 | 0.22 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr6_-_12722360 | 0.22 |
ENSDART00000090174
|
dock9b
|
dedicator of cytokinesis 9b |
| chr25_+_36045072 | 0.22 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr15_+_9327252 | 0.22 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr22_-_18179214 | 0.22 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr4_-_23858900 | 0.21 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr8_-_24965308 | 0.21 |
ENSDART00000087293
ENSDART00000146805 |
kcnc4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr10_+_24468922 | 0.21 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr13_+_2894536 | 0.21 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr10_-_24371312 | 0.21 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr19_-_33087246 | 0.21 |
ENSDART00000052078
|
klhl38a
|
kelch-like family member 38a |
| chr16_+_11151699 | 0.21 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr10_-_25069155 | 0.21 |
ENSDART00000078226
ENSDART00000181941 |
mtnr1bb
|
melatonin receptor 1Bb |
| chr20_+_46560258 | 0.21 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr4_+_20263097 | 0.21 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr6_+_29217392 | 0.20 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr8_+_50727220 | 0.20 |
ENSDART00000127062
|
egr3
|
early growth response 3 |
| chr3_+_22935183 | 0.20 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr19_+_42770041 | 0.20 |
ENSDART00000150930
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr19_+_7001170 | 0.20 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr7_+_6652967 | 0.19 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr21_-_24865454 | 0.19 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr6_-_8865217 | 0.19 |
ENSDART00000151443
|
tmeff2b
|
transmembrane protein with EGF-like and two follistatin-like domains 2b |
| chr16_-_42750295 | 0.19 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr18_+_19820008 | 0.19 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr8_+_48484455 | 0.19 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr17_+_8292892 | 0.18 |
ENSDART00000125728
|
si:ch211-236p5.3
|
si:ch211-236p5.3 |
| chr15_+_33989181 | 0.18 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr20_-_42439896 | 0.18 |
ENSDART00000061049
|
vgll2a
|
vestigial-like family member 2a |
| chr19_-_32888758 | 0.18 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr3_-_23596532 | 0.18 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr3_+_20156956 | 0.17 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr3_+_36515376 | 0.17 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr20_-_18915376 | 0.17 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr4_+_12113105 | 0.17 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr11_-_17713987 | 0.17 |
ENSDART00000090401
|
fam19a4b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4b |
| chr13_+_43400443 | 0.17 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr5_+_34622320 | 0.17 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr9_-_8454060 | 0.17 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr4_-_18635005 | 0.17 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr7_+_27253063 | 0.17 |
ENSDART00000191138
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_+_24251367 | 0.17 |
ENSDART00000173642
|
svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr4_-_20081621 | 0.16 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr6_-_35032792 | 0.16 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr23_-_26880623 | 0.16 |
ENSDART00000038491
|
adcy6b
|
adenylate cyclase 6b |
| chr23_+_13528550 | 0.16 |
ENSDART00000099903
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr22_+_16308450 | 0.16 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr6_-_8865543 | 0.16 |
ENSDART00000042115
|
tmeff2b
|
transmembrane protein with EGF-like and two follistatin-like domains 2b |
| chr15_-_29387446 | 0.16 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr24_+_26276805 | 0.16 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr19_+_44039849 | 0.16 |
ENSDART00000086040
|
lrrc14b
|
leucine rich repeat containing 14B |
| chr3_-_23596809 | 0.16 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr3_-_28075756 | 0.16 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr6_+_4872883 | 0.16 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr9_+_38168012 | 0.15 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr4_-_17823424 | 0.15 |
ENSDART00000024822
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr20_-_18731268 | 0.15 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr5_-_60159116 | 0.15 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr25_-_19655820 | 0.15 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr25_-_35542739 | 0.15 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr17_+_12075805 | 0.15 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr1_-_17587552 | 0.15 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr11_-_36963988 | 0.14 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr13_+_18545819 | 0.14 |
ENSDART00000101859
ENSDART00000110197 ENSDART00000136064 |
zgc:154058
|
zgc:154058 |
| chr8_+_36948256 | 0.14 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr18_-_7948188 | 0.14 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr14_-_9355177 | 0.13 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr7_-_48263516 | 0.13 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr21_-_38618540 | 0.13 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_+_20471315 | 0.13 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr8_+_7033049 | 0.13 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr2_+_4208323 | 0.13 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.6 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.5 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.1 | 0.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.5 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.7 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.1 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.8 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.9 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.0 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.0 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.6 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.0 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.2 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.2 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 0.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |