Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
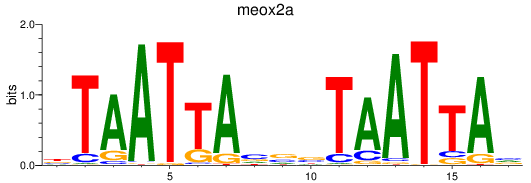
Results for meox2a
Z-value: 1.05
Transcription factors associated with meox2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox2a
|
ENSDARG00000040911 | mesenchyme homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox2a | dr11_v1_chr15_-_34468599_34468599 | 0.80 | 1.0e-01 | Click! |
Activity profile of meox2a motif
Sorted Z-values of meox2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31267268 | 1.21 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr11_-_2131280 | 0.93 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr23_-_27571667 | 0.84 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr5_+_32222303 | 0.79 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr11_-_1509773 | 0.55 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr6_+_55032439 | 0.54 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr2_-_30324610 | 0.53 |
ENSDART00000185422
|
jph1b
|
junctophilin 1b |
| chr23_+_26026383 | 0.45 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_32150078 | 0.41 |
ENSDART00000134934
ENSDART00000186410 ENSDART00000181780 |
pag1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr7_+_35245607 | 0.40 |
ENSDART00000193422
ENSDART00000173888 |
amfrb
|
autocrine motility factor receptor b |
| chr12_-_26407092 | 0.39 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr4_+_9669717 | 0.37 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr25_+_20077225 | 0.36 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr2_+_36004381 | 0.35 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr16_+_12836143 | 0.31 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr2_+_24188502 | 0.30 |
ENSDART00000181955
ENSDART00000191706 ENSDART00000125909 |
map4l
|
microtubule associated protein 4 like |
| chr24_+_24831727 | 0.28 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr12_+_30586599 | 0.27 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr25_+_20089986 | 0.27 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr19_+_5072918 | 0.26 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr2_+_50862527 | 0.26 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr22_-_10459880 | 0.25 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr14_+_924876 | 0.24 |
ENSDART00000183908
|
myoz3a
|
myozenin 3a |
| chr15_-_3252727 | 0.24 |
ENSDART00000131173
|
stoml3a
|
stomatin (EPB72)-like 3a |
| chr2_+_57801960 | 0.23 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr14_-_3032016 | 0.23 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr22_+_15438872 | 0.23 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr3_-_39152478 | 0.22 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr21_+_28958471 | 0.21 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_-_5669122 | 0.21 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr20_+_1960092 | 0.21 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr4_-_17793152 | 0.21 |
ENSDART00000134080
|
mybpc1
|
myosin binding protein C, slow type |
| chr19_-_29887629 | 0.20 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr16_-_24605969 | 0.19 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr6_-_11768198 | 0.19 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr3_-_32169754 | 0.18 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr14_+_22113331 | 0.18 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr13_+_43247936 | 0.18 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr5_+_23622177 | 0.18 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr12_-_3753131 | 0.18 |
ENSDART00000129668
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr17_-_49978986 | 0.17 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr1_+_31110817 | 0.17 |
ENSDART00000137863
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr5_+_7279104 | 0.16 |
ENSDART00000190014
|
si:ch73-72b7.1
|
si:ch73-72b7.1 |
| chr18_-_1258777 | 0.16 |
ENSDART00000077106
ENSDART00000129065 |
ugt5f1
|
UDP glucuronosyltransferase 5 family, polypeptide F1 |
| chr9_-_8661436 | 0.16 |
ENSDART00000130442
|
col4a1
|
collagen, type IV, alpha 1 |
| chr1_-_12278056 | 0.16 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr7_-_5162292 | 0.16 |
ENSDART00000084218
|
zgc:195075
|
zgc:195075 |
| chr19_-_28360033 | 0.16 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr9_-_31596016 | 0.15 |
ENSDART00000142289
|
nalcn
|
sodium leak channel, non-selective |
| chr23_-_11870962 | 0.15 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr5_-_37117778 | 0.15 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr19_-_2861444 | 0.14 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr5_-_52277643 | 0.14 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr1_+_17593392 | 0.14 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr4_+_25223050 | 0.14 |
ENSDART00000138121
|
itih5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr18_+_7286788 | 0.13 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr5_+_20147830 | 0.13 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr2_+_50608099 | 0.13 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr24_-_7321928 | 0.13 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr25_+_20091021 | 0.13 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr2_-_2957970 | 0.13 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr23_+_45538932 | 0.13 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr7_-_51461649 | 0.13 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr6_-_55585423 | 0.13 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr6_-_19271210 | 0.13 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr4_-_77506362 | 0.13 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr18_+_5137241 | 0.13 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr8_+_49778486 | 0.13 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr9_-_52847881 | 0.12 |
ENSDART00000166407
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr8_+_36942262 | 0.12 |
ENSDART00000188173
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr2_+_34572690 | 0.12 |
ENSDART00000077216
|
astn1
|
astrotactin 1 |
| chr16_-_24642814 | 0.12 |
ENSDART00000153987
ENSDART00000154319 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr24_-_29963858 | 0.11 |
ENSDART00000183442
|
CR352310.1
|
|
| chr19_-_5865766 | 0.11 |
ENSDART00000191007
|
LO018585.1
|
|
| chr17_-_16965809 | 0.11 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr16_-_43344859 | 0.11 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr17_+_15535501 | 0.11 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr21_+_33187992 | 0.11 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr9_-_51323545 | 0.11 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr13_+_29778610 | 0.11 |
ENSDART00000132004
|
pax2a
|
paired box 2a |
| chr2_-_31521764 | 0.11 |
ENSDART00000140523
|
mrc1b
|
mannose receptor, C type 1b |
| chr1_+_45085194 | 0.11 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr7_+_14632157 | 0.10 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr24_+_33802528 | 0.10 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr9_+_2020667 | 0.10 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr8_-_14121634 | 0.10 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr11_+_25129013 | 0.10 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr22_-_881725 | 0.10 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr6_+_13726844 | 0.10 |
ENSDART00000055833
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr5_+_6672870 | 0.10 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr5_-_68093169 | 0.10 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr17_+_3379673 | 0.09 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr16_+_5184402 | 0.09 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr3_+_59784632 | 0.09 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr6_-_33023745 | 0.09 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr16_-_27174373 | 0.09 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr20_-_40755614 | 0.09 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr21_-_11654422 | 0.09 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr5_+_25952340 | 0.09 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr20_-_42102416 | 0.09 |
ENSDART00000186378
ENSDART00000188253 ENSDART00000186458 |
slc35f1
|
solute carrier family 35, member F1 |
| chr24_+_14937205 | 0.09 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr3_+_62271320 | 0.09 |
ENSDART00000132197
|
BX470259.2
|
|
| chr15_+_44366556 | 0.08 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr20_+_26966725 | 0.08 |
ENSDART00000029781
|
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr12_-_28570989 | 0.08 |
ENSDART00000008010
|
pdk2a
|
pyruvate dehydrogenase kinase, isozyme 2a |
| chr1_-_22512063 | 0.08 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr20_+_4060839 | 0.08 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr6_-_32025225 | 0.08 |
ENSDART00000006417
|
pgm1
|
phosphoglucomutase 1 |
| chr23_+_16620801 | 0.08 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr8_-_14080534 | 0.08 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr25_-_8030113 | 0.08 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr16_-_41944765 | 0.08 |
ENSDART00000142648
|
gramd1a
|
GRAM domain containing 1A |
| chr5_+_51026563 | 0.08 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr23_-_15284757 | 0.08 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr10_+_35952532 | 0.07 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr23_+_36106790 | 0.07 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr15_-_26844591 | 0.07 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr19_+_310494 | 0.07 |
ENSDART00000053638
|
ctss1
|
cathepsin S, ortholog 1 |
| chr12_+_18681477 | 0.07 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr8_-_979735 | 0.07 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr21_-_26071142 | 0.07 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr10_-_44508249 | 0.07 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr21_-_13123176 | 0.07 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr3_-_19091024 | 0.07 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr5_+_20035284 | 0.07 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr21_-_43015383 | 0.07 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr22_+_33131891 | 0.07 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr6_+_18423402 | 0.07 |
ENSDART00000159747
|
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr8_-_13972626 | 0.07 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr14_+_25465346 | 0.07 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr24_+_23742690 | 0.07 |
ENSDART00000130162
|
TCF24
|
transcription factor 24 |
| chr20_-_1378514 | 0.07 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr18_+_3332999 | 0.06 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr10_+_5203532 | 0.06 |
ENSDART00000165018
|
cdc42se2
|
CDC42 small effector 2 |
| chr18_-_48558420 | 0.06 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr16_-_6849754 | 0.06 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr17_+_21902058 | 0.06 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr14_-_1646360 | 0.06 |
ENSDART00000186528
|
LO018106.2
|
|
| chr2_-_144393 | 0.06 |
ENSDART00000156008
|
adcy1b
|
adenylate cyclase 1b |
| chr16_+_23598908 | 0.06 |
ENSDART00000131627
|
kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr21_-_26071773 | 0.06 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr25_-_10565006 | 0.06 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr5_+_59392183 | 0.06 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr10_+_21576909 | 0.06 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr5_+_29159777 | 0.06 |
ENSDART00000174702
ENSDART00000037891 |
dpp7
|
dipeptidyl-peptidase 7 |
| chr4_-_48957129 | 0.06 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr20_-_47051996 | 0.06 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr12_+_13652361 | 0.06 |
ENSDART00000182757
ENSDART00000152689 |
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr10_+_26926654 | 0.06 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr12_-_47580562 | 0.06 |
ENSDART00000153431
|
rgs7b
|
regulator of G protein signaling 7b |
| chr15_-_44077937 | 0.06 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr24_-_20208474 | 0.06 |
ENSDART00000139329
|
cry-dash
|
cryptochrome DASH |
| chr25_+_26895394 | 0.06 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr21_-_11655010 | 0.06 |
ENSDART00000144370
ENSDART00000139814 ENSDART00000139289 |
cast
|
calpastatin |
| chr6_+_49412754 | 0.06 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr23_-_33738570 | 0.06 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr6_-_52723901 | 0.06 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr9_+_49712868 | 0.06 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr1_+_25801648 | 0.06 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr12_+_17504559 | 0.06 |
ENSDART00000020628
|
cyth3a
|
cytohesin 3a |
| chr15_+_6114109 | 0.05 |
ENSDART00000184937
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr5_-_67783593 | 0.05 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr19_-_5805923 | 0.05 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr21_-_29991098 | 0.05 |
ENSDART00000155772
|
si:ch73-190f9.4
|
si:ch73-190f9.4 |
| chr4_-_119689 | 0.05 |
ENSDART00000161055
|
dusp16
|
dual specificity phosphatase 16 |
| chr17_+_26611929 | 0.05 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr25_-_8030425 | 0.05 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr9_+_46644633 | 0.05 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr22_-_5553007 | 0.05 |
ENSDART00000165269
|
zgc:171601
|
zgc:171601 |
| chr7_-_46777876 | 0.05 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr21_+_1381276 | 0.05 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr8_+_2438638 | 0.05 |
ENSDART00000141263
|
ENKD1
|
si:ch211-220d9.3 |
| chr2_+_56937548 | 0.05 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr25_-_4713461 | 0.05 |
ENSDART00000155302
|
drd4a
|
dopamine receptor D4a |
| chr3_+_52275691 | 0.05 |
ENSDART00000190109
|
BX511132.2
|
|
| chr9_-_52920305 | 0.05 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr1_-_45920632 | 0.05 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr3_+_62327332 | 0.05 |
ENSDART00000158414
|
BX470259.1
|
|
| chr7_+_65261576 | 0.05 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr2_-_14390627 | 0.05 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr12_-_23320266 | 0.05 |
ENSDART00000181711
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr7_+_25858380 | 0.04 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr25_-_19594065 | 0.04 |
ENSDART00000146578
ENSDART00000003715 |
actr6
|
ARP6 actin related protein 6 homolog |
| chr10_-_2524917 | 0.04 |
ENSDART00000188642
|
CU856539.1
|
|
| chr13_-_48756036 | 0.04 |
ENSDART00000035018
ENSDART00000132895 |
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr9_-_34882516 | 0.04 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr7_+_8456999 | 0.04 |
ENSDART00000172880
|
jac4
|
jacalin 4 |
| chr14_-_26094172 | 0.04 |
ENSDART00000150131
|
mtnr1al
|
melatonin receptor type 1A like |
| chr16_+_20161805 | 0.04 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr13_-_35808904 | 0.04 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr3_-_43954971 | 0.04 |
ENSDART00000167159
ENSDART00000189315 |
ubfd1
|
ubiquitin family domain containing 1 |
| chr10_+_8730988 | 0.04 |
ENSDART00000145596
|
itga1
|
integrin, alpha 1 |
| chr15_+_4969128 | 0.04 |
ENSDART00000062856
|
rnf169
|
ring finger protein 169 |
| chr16_+_11834516 | 0.04 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr20_-_18382708 | 0.04 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr19_-_40192249 | 0.04 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr5_+_69950882 | 0.04 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr13_+_11828516 | 0.04 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr18_+_11506561 | 0.04 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr19_+_42227400 | 0.04 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 2.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) sensory perception of temperature stimulus(GO:0050951) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.1 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.0 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.0 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.6 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |