Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
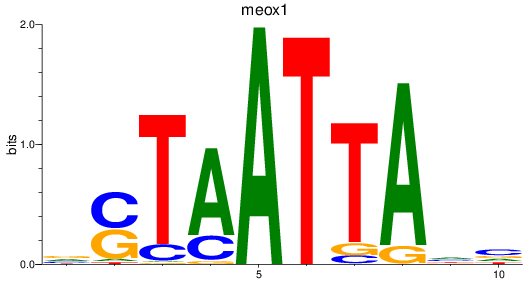
Results for meox1
Z-value: 0.72
Transcription factors associated with meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meox1
|
ENSDARG00000007891 | mesenchyme homeobox 1 |
|
meox1
|
ENSDARG00000115382 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meox1 | dr11_v1_chr12_+_27462225_27462225 | -0.13 | 8.3e-01 | Click! |
Activity profile of meox1 motif
Sorted Z-values of meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10175898 | 0.51 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr10_-_21955231 | 0.37 |
ENSDART00000183695
|
FO744833.2
|
|
| chr1_-_23308225 | 0.36 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr11_+_44135351 | 0.35 |
ENSDART00000182914
|
FO704721.1
|
|
| chr3_-_61162750 | 0.34 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr3_-_31254979 | 0.34 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr12_+_22580579 | 0.34 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr20_-_37813863 | 0.33 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr2_+_15048410 | 0.32 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr20_-_9462433 | 0.31 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr11_-_25418856 | 0.29 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr3_+_13624815 | 0.29 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr25_+_10410620 | 0.28 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr20_-_9436521 | 0.26 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr8_-_31107537 | 0.25 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr22_-_13350240 | 0.25 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr7_-_25895189 | 0.24 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr6_-_43283122 | 0.22 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr16_+_24684107 | 0.22 |
ENSDART00000183920
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr17_+_12865746 | 0.22 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr23_-_18130264 | 0.22 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr9_+_43799829 | 0.22 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr24_-_39518599 | 0.21 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr21_+_26726936 | 0.21 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_-_31254379 | 0.21 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr23_+_20110086 | 0.20 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr21_+_11244068 | 0.20 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr20_+_46620374 | 0.20 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr6_-_40581376 | 0.20 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr16_-_28878080 | 0.19 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr15_+_21262917 | 0.19 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr5_+_38263240 | 0.19 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr20_+_46513651 | 0.17 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr12_+_20352400 | 0.17 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr19_+_7735157 | 0.17 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr1_-_44161417 | 0.17 |
ENSDART00000083213
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr3_+_25087605 | 0.17 |
ENSDART00000138578
|
st13
|
ST13, Hsp70 interacting protein |
| chr4_+_9177997 | 0.17 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr4_+_11464255 | 0.17 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr23_-_31913069 | 0.17 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_-_26490186 | 0.17 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr24_-_41320037 | 0.17 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr22_-_11137268 | 0.17 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr15_+_21276735 | 0.16 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr23_-_36446307 | 0.16 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr25_+_5035343 | 0.16 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr22_+_5176693 | 0.16 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr17_+_16046314 | 0.16 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_+_45364849 | 0.16 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr8_-_30338872 | 0.16 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr19_-_32641725 | 0.16 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr19_+_12406583 | 0.16 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr19_-_42607451 | 0.15 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr1_-_50859053 | 0.15 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr17_+_30545895 | 0.15 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr20_-_23226453 | 0.15 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr23_+_36095260 | 0.15 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr22_+_5176255 | 0.15 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr3_+_45365098 | 0.14 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr23_-_35790235 | 0.14 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr21_-_7035599 | 0.14 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr12_-_46112892 | 0.14 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr14_-_14640401 | 0.14 |
ENSDART00000168027
ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr24_+_12133814 | 0.14 |
ENSDART00000158562
ENSDART00000159029 ENSDART00000168248 |
lztfl1
|
leucine zipper transcription factor-like 1 |
| chr7_+_20110336 | 0.14 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr14_+_23717165 | 0.13 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_+_6576940 | 0.13 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr19_-_5103141 | 0.13 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr20_-_45060241 | 0.13 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr9_+_38372216 | 0.13 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr24_+_16547035 | 0.13 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr17_+_21902058 | 0.13 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr11_-_23687158 | 0.13 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr25_+_35553542 | 0.13 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr18_+_8833251 | 0.13 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr4_+_77943184 | 0.12 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_-_25649079 | 0.12 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr18_+_2228737 | 0.12 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr6_-_12275836 | 0.12 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr17_-_31695217 | 0.12 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr5_+_27897504 | 0.12 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr19_-_205104 | 0.12 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr5_-_30074332 | 0.12 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr2_-_23172708 | 0.12 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr17_+_16046132 | 0.12 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr7_+_2236317 | 0.11 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr2_-_28671139 | 0.11 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr15_-_23376541 | 0.11 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr11_+_14284866 | 0.11 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr3_-_31079186 | 0.11 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr15_-_36533322 | 0.11 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr8_-_21142550 | 0.11 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr11_-_45138857 | 0.11 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_39120306 | 0.11 |
ENSDART00000155746
ENSDART00000154158 |
cdc42
|
cell division cycle 42 |
| chr13_+_35339182 | 0.11 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_+_43347966 | 0.11 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr7_+_23292133 | 0.10 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr13_+_27232848 | 0.10 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr7_-_45999333 | 0.10 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr23_+_31912882 | 0.10 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr22_-_21897203 | 0.10 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr20_+_29209615 | 0.10 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_20931197 | 0.10 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_+_23712911 | 0.10 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr8_-_30204650 | 0.09 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr11_+_25129013 | 0.09 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr22_-_13042992 | 0.09 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr10_+_16036246 | 0.09 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr11_-_29768054 | 0.09 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr23_+_31913292 | 0.09 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr24_+_39518774 | 0.09 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr2_+_32826235 | 0.09 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr8_-_24252933 | 0.09 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr20_-_22476255 | 0.09 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr15_-_9272328 | 0.09 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr17_-_37395460 | 0.09 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr12_-_11560794 | 0.09 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr1_-_10647307 | 0.08 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr3_+_27786601 | 0.08 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr15_-_1038193 | 0.08 |
ENSDART00000159462
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr10_+_2582254 | 0.08 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr9_-_38021889 | 0.08 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr3_+_32443395 | 0.08 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr24_+_36204028 | 0.08 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr15_-_4528326 | 0.08 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr12_-_33972798 | 0.08 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr18_+_15758375 | 0.08 |
ENSDART00000137554
|
si:ch211-219a15.4
|
si:ch211-219a15.4 |
| chr9_-_3934963 | 0.08 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr8_-_52562672 | 0.08 |
ENSDART00000159333
ENSDART00000159974 |
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr8_-_25034411 | 0.08 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr25_-_6049339 | 0.08 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr14_-_24081929 | 0.08 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr14_+_34971554 | 0.08 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr6_+_3280939 | 0.08 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr19_+_40856807 | 0.08 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr23_-_900795 | 0.08 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr2_-_26720854 | 0.07 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr19_+_15521997 | 0.07 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr16_+_28994709 | 0.07 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chrM_+_9052 | 0.07 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr19_+_40856534 | 0.07 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr7_-_30174882 | 0.07 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr5_+_40837539 | 0.07 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr15_+_28410664 | 0.07 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr22_-_881725 | 0.07 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr13_+_30903816 | 0.07 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr19_+_47476908 | 0.07 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr6_-_44044385 | 0.07 |
ENSDART00000075497
|
rybpb
|
RING1 and YY1 binding protein b |
| chr8_+_52314542 | 0.07 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr19_-_5103313 | 0.06 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_-_31913231 | 0.06 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr1_+_51191049 | 0.06 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_-_23117078 | 0.06 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr2_+_26288301 | 0.06 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr4_-_17629444 | 0.06 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr3_-_26805455 | 0.06 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr3_-_16719244 | 0.06 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_-_14813029 | 0.06 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr5_-_68093169 | 0.06 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr6_-_35472923 | 0.06 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr2_+_57848844 | 0.06 |
ENSDART00000037279
|
plekhj1
|
pleckstrin homology domain containing, family J member 1 |
| chr20_-_20930926 | 0.06 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr24_-_34680956 | 0.06 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr16_-_26855936 | 0.05 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr10_-_2971407 | 0.05 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr20_+_29209767 | 0.05 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr12_+_30367371 | 0.05 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr3_+_22442445 | 0.05 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chrM_+_9735 | 0.05 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr10_+_16584382 | 0.05 |
ENSDART00000112039
|
CR790388.1
|
|
| chr3_-_26806032 | 0.05 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_1313418 | 0.05 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr17_+_12813316 | 0.05 |
ENSDART00000155629
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr11_+_31864921 | 0.05 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr6_+_54576520 | 0.05 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr10_+_3049636 | 0.05 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr21_-_25565392 | 0.05 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr18_+_8402129 | 0.05 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr9_-_43538328 | 0.04 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr25_+_32755485 | 0.04 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr3_+_1637358 | 0.04 |
ENSDART00000180266
|
CR394546.5
|
|
| chr20_+_29209926 | 0.04 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_-_18744511 | 0.04 |
ENSDART00000145539
|
zgc:113333
|
zgc:113333 |
| chr18_-_2433011 | 0.04 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr1_-_19402802 | 0.04 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr4_+_25917915 | 0.04 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr15_+_1796313 | 0.04 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr17_-_21162821 | 0.04 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr6_-_24392426 | 0.04 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr3_+_34919810 | 0.04 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr24_-_40860603 | 0.04 |
ENSDART00000188032
|
CU633479.7
|
|
| chr17_-_20118145 | 0.04 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr20_-_46362606 | 0.04 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr6_+_21001264 | 0.04 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr22_+_17205608 | 0.04 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr4_-_16124417 | 0.04 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr24_+_22485710 | 0.04 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr7_-_64589920 | 0.04 |
ENSDART00000172619
ENSDART00000184113 |
CR387919.1
|
|
| chr16_-_21038015 | 0.03 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr2_-_27619954 | 0.03 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.5 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.2 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.2 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.1 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.0 | 0.0 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |