Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
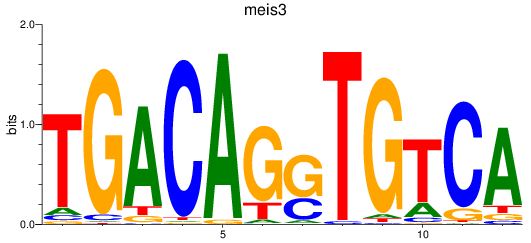
Results for meis3
Z-value: 0.54
Transcription factors associated with meis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis3
|
ENSDARG00000002795 | myeloid ecotropic viral integration site 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis3 | dr11_v1_chr15_-_6863317_6863317 | 0.62 | 2.6e-01 | Click! |
Activity profile of meis3 motif
Sorted Z-values of meis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10177442 | 0.59 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr15_-_29598679 | 0.38 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr6_+_3827751 | 0.33 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr15_-_29598444 | 0.31 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr21_+_45733871 | 0.30 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr10_-_39011514 | 0.30 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr6_-_14146979 | 0.27 |
ENSDART00000089564
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr18_+_36769758 | 0.27 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr6_+_48041759 | 0.25 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr8_+_26329482 | 0.25 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr5_+_24245682 | 0.25 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr15_-_4415917 | 0.24 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr8_+_25959940 | 0.23 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr23_-_15216654 | 0.22 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr6_-_39631164 | 0.22 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr17_+_33495194 | 0.21 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr10_+_26961965 | 0.20 |
ENSDART00000089144
|
vps51
|
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr19_+_24872159 | 0.19 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr2_+_6127593 | 0.19 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr8_-_27515540 | 0.19 |
ENSDART00000146132
|
si:ch211-254n4.3
|
si:ch211-254n4.3 |
| chr9_-_28399071 | 0.18 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr6_+_9107063 | 0.18 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr19_-_30565122 | 0.17 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
| chr10_+_26667475 | 0.17 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr5_-_46980651 | 0.16 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr3_-_21118969 | 0.16 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr22_-_22231720 | 0.15 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr5_+_37091626 | 0.14 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr5_+_23118470 | 0.14 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr6_+_33537267 | 0.13 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr23_+_23182037 | 0.13 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr6_-_13408680 | 0.13 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr2_-_44183613 | 0.13 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr2_+_47623202 | 0.13 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr16_-_11986321 | 0.12 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr2_-_44183451 | 0.12 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr11_-_18799827 | 0.12 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr18_+_36770166 | 0.12 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr7_-_19369002 | 0.12 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr4_+_20812900 | 0.12 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr24_-_30091937 | 0.12 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr23_-_7125494 | 0.12 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr14_-_26436951 | 0.12 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr19_+_42693855 | 0.11 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr14_-_17305657 | 0.10 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr7_-_18881358 | 0.10 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr6_-_30210378 | 0.10 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr8_-_1838315 | 0.10 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr23_-_20126257 | 0.10 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr5_-_15283509 | 0.10 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr9_-_29844596 | 0.09 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr5_-_43682930 | 0.09 |
ENSDART00000075017
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr7_-_67214972 | 0.09 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr12_+_36413886 | 0.09 |
ENSDART00000126325
|
si:ch211-250n8.1
|
si:ch211-250n8.1 |
| chr19_-_42588510 | 0.09 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr20_-_22798794 | 0.09 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr2_-_32558795 | 0.08 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr19_+_20713424 | 0.08 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr21_+_30194904 | 0.08 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr8_-_32506569 | 0.08 |
ENSDART00000061792
|
RFESD (1 of many)
|
Rieske Fe-S domain containing |
| chr5_-_32396929 | 0.08 |
ENSDART00000023977
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr6_+_9952678 | 0.08 |
ENSDART00000019325
|
cyp20a1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr22_+_30335936 | 0.08 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr14_-_9355177 | 0.07 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr10_-_32877348 | 0.07 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr23_+_25172682 | 0.07 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr8_-_7444155 | 0.07 |
ENSDART00000148993
|
hdac6
|
histone deacetylase 6 |
| chr24_+_17349177 | 0.07 |
ENSDART00000176338
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr10_-_33379850 | 0.07 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr13_+_22295905 | 0.06 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr25_-_7753207 | 0.06 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr1_+_34496855 | 0.06 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr7_+_6879534 | 0.06 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr25_-_10610961 | 0.06 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr8_+_144154 | 0.05 |
ENSDART00000164099
|
snx2
|
sorting nexin 2 |
| chr4_-_25908871 | 0.05 |
ENSDART00000066949
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_-_45308394 | 0.05 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr24_+_12989727 | 0.05 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr13_+_30169681 | 0.04 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr6_+_15762647 | 0.04 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr23_-_10696626 | 0.04 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr20_+_44311448 | 0.04 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr17_-_43552894 | 0.04 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr17_-_15667198 | 0.04 |
ENSDART00000142972
ENSDART00000132571 ENSDART00000189936 |
manea
|
mannosidase, endo-alpha |
| chr16_-_29146624 | 0.04 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr23_+_25172976 | 0.04 |
ENSDART00000140789
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr8_-_25569920 | 0.03 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr18_+_660578 | 0.03 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr10_+_4964463 | 0.03 |
ENSDART00000109882
|
lyrm7
|
LYR motif containing 7 |
| chr25_-_22191733 | 0.03 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr7_-_26306546 | 0.03 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr2_+_45696743 | 0.03 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr5_-_37341044 | 0.02 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr2_+_36112273 | 0.02 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr20_-_26467307 | 0.02 |
ENSDART00000078072
ENSDART00000158213 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr15_-_23442891 | 0.02 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr8_-_4508248 | 0.02 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr14_-_51014292 | 0.01 |
ENSDART00000029797
|
faf2
|
Fas associated factor family member 2 |
| chr8_-_18225968 | 0.01 |
ENSDART00000135504
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr19_+_34742706 | 0.01 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr23_-_33680265 | 0.01 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr5_+_57726425 | 0.01 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr20_-_26421112 | 0.01 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr20_-_26420939 | 0.01 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr2_-_39090258 | 0.01 |
ENSDART00000134270
ENSDART00000145268 |
NMNAT3
|
si:ch73-170l17.1 |
| chr23_+_21663631 | 0.01 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr7_-_30174882 | 0.00 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr20_-_23171430 | 0.00 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr23_-_33679579 | 0.00 |
ENSDART00000188674
|
tfcp2
|
transcription factor CP2 |
| chr25_+_19670273 | 0.00 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr17_+_7595356 | 0.00 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.2 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |