Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
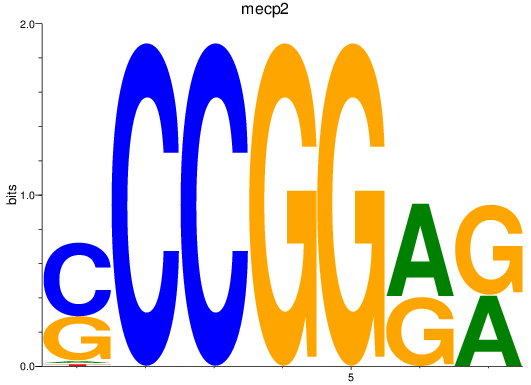
Results for mecp2
Z-value: 2.42
Transcription factors associated with mecp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecp2
|
ENSDARG00000014218 | methyl CpG binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecp2 | dr11_v1_chr8_-_7637626_7637640 | 0.66 | 2.2e-01 | Click! |
Activity profile of mecp2 motif
Sorted Z-values of mecp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_13685047 | 2.28 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr8_+_16004154 | 2.18 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr22_+_11535131 | 2.14 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr9_-_31278048 | 2.05 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr8_-_25422186 | 2.01 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr12_-_29624638 | 1.85 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr16_+_32559821 | 1.79 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr3_+_31396149 | 1.78 |
ENSDART00000151423
ENSDART00000193580 |
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr5_+_64739762 | 1.76 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr19_+_232536 | 1.66 |
ENSDART00000137880
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr4_+_19534833 | 1.65 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr1_+_31864404 | 1.61 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr8_+_31119548 | 1.59 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr19_+_5072918 | 1.57 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr8_+_16004551 | 1.56 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr15_+_39096736 | 1.52 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr1_+_12766351 | 1.43 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr2_+_27491066 | 1.42 |
ENSDART00000078305
|
zswim5
|
zinc finger, SWIM-type containing 5 |
| chr20_-_45661049 | 1.42 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr17_-_6759006 | 1.41 |
ENSDART00000184692
ENSDART00000180530 |
vsnl1b
|
visinin-like 1b |
| chr25_-_19443421 | 1.38 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr3_+_35005062 | 1.38 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr19_-_47587719 | 1.37 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr23_+_2361184 | 1.37 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr21_-_10446405 | 1.33 |
ENSDART00000167948
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr11_-_762721 | 1.30 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr18_-_46881108 | 1.28 |
ENSDART00000190084
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr17_-_19022990 | 1.28 |
ENSDART00000154186
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_-_1360495 | 1.24 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr6_+_41554794 | 1.24 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr20_-_30035326 | 1.22 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr9_+_24159725 | 1.22 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr1_-_40994259 | 1.20 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr19_+_56351 | 1.19 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr8_+_47897734 | 1.18 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr20_+_22666548 | 1.15 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr10_-_22845485 | 1.15 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr25_+_8063455 | 1.13 |
ENSDART00000073919
|
kcnc1b
|
potassium voltage-gated channel, Shaw-related subfamily, member 1b |
| chr3_+_46724528 | 1.11 |
ENSDART00000181358
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr12_+_33038757 | 1.09 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr3_-_27880229 | 1.09 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr9_+_17787864 | 1.08 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr16_-_29458806 | 1.07 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr1_-_20911297 | 1.07 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr20_+_32523576 | 1.06 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr13_-_40120252 | 1.05 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr17_+_28883353 | 1.04 |
ENSDART00000110322
|
prkd1
|
protein kinase D1 |
| chr14_-_2355833 | 1.04 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr13_-_36911118 | 1.04 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr7_+_19552381 | 1.04 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_+_34572690 | 1.03 |
ENSDART00000077216
|
astn1
|
astrotactin 1 |
| chr4_+_23223881 | 1.03 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr8_+_23165749 | 1.00 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr14_-_2050057 | 1.00 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr22_-_13544244 | 1.00 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr9_-_18877597 | 0.99 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr16_-_31188715 | 0.99 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr14_-_9199968 | 0.98 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr2_-_50372467 | 0.98 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr18_+_5917625 | 0.94 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr20_-_47704973 | 0.93 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr1_+_54908895 | 0.92 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr16_+_34528409 | 0.90 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr1_+_25783801 | 0.90 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr11_-_27917730 | 0.90 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr17_-_28198099 | 0.89 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr4_-_12102025 | 0.89 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr21_-_42100471 | 0.89 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr15_+_28685625 | 0.89 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr5_-_28606916 | 0.88 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr25_-_34156152 | 0.88 |
ENSDART00000125036
|
foxb1a
|
forkhead box B1a |
| chr19_-_9503473 | 0.87 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr17_+_28882977 | 0.86 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr10_+_39476600 | 0.86 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr24_+_5208171 | 0.85 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr8_+_21146262 | 0.84 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr20_+_474288 | 0.84 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr15_+_45994123 | 0.81 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr15_+_40188076 | 0.81 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr11_-_37589293 | 0.81 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr3_+_17806213 | 0.81 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr20_+_52389858 | 0.81 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr20_-_47732703 | 0.81 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr16_-_12914288 | 0.80 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr6_-_21988375 | 0.80 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr18_+_28564640 | 0.80 |
ENSDART00000016983
|
spon1a
|
spondin 1a |
| chr3_-_5664123 | 0.80 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr8_+_48966165 | 0.79 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr24_+_24461558 | 0.78 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr5_-_29488245 | 0.78 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr15_+_28685892 | 0.78 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr19_+_32979331 | 0.77 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr6_+_39222598 | 0.77 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr15_-_18138607 | 0.77 |
ENSDART00000176690
|
CR385077.1
|
|
| chr9_-_55790458 | 0.76 |
ENSDART00000161247
|
sowahca
|
sosondowah ankyrin repeat domain family Ca |
| chr20_-_31905968 | 0.76 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr8_-_30979494 | 0.75 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr8_-_22739757 | 0.74 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr13_-_36545258 | 0.71 |
ENSDART00000186171
|
FP103009.1
|
|
| chr18_+_45228691 | 0.71 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr6_+_58406014 | 0.70 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr4_+_20263097 | 0.70 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr15_-_47895200 | 0.70 |
ENSDART00000027060
|
DMWD
|
zmp:0000000529 |
| chr14_+_5936996 | 0.69 |
ENSDART00000097144
ENSDART00000126777 |
kctd8
|
potassium channel tetramerization domain containing 8 |
| chr13_-_36184476 | 0.69 |
ENSDART00000057185
|
map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr16_+_1353894 | 0.68 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr14_-_27289042 | 0.68 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr23_+_28128453 | 0.68 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr2_-_37401600 | 0.67 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr19_+_37458610 | 0.67 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr21_+_28728030 | 0.65 |
ENSDART00000097307
|
puraa
|
purine-rich element binding protein Aa |
| chr15_-_45110011 | 0.65 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr25_-_20381107 | 0.65 |
ENSDART00000067454
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr5_+_59397739 | 0.65 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr2_-_32501501 | 0.65 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr17_+_52300018 | 0.64 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr7_-_24699985 | 0.63 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr2_+_39108339 | 0.63 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr10_-_43392667 | 0.63 |
ENSDART00000183033
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr17_+_43629008 | 0.63 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr14_-_2196267 | 0.62 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr7_+_1442059 | 0.62 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr5_+_26079178 | 0.60 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr25_-_12203952 | 0.60 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr25_+_17589906 | 0.60 |
ENSDART00000167750
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr21_-_37790727 | 0.60 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr15_+_23208042 | 0.60 |
ENSDART00000006085
|
cbl
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr15_+_2559875 | 0.59 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr7_-_26389908 | 0.59 |
ENSDART00000109268
|
si:dkey-3k24.5
|
si:dkey-3k24.5 |
| chr19_-_32914227 | 0.59 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr6_+_48664275 | 0.59 |
ENSDART00000161184
|
FAM19A3
|
si:dkey-238f9.1 |
| chr3_+_13637383 | 0.58 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr3_-_35602233 | 0.58 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr2_+_50391331 | 0.58 |
ENSDART00000098108
|
viml
|
vimentin like |
| chr25_-_20381271 | 0.58 |
ENSDART00000142665
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr2_+_40294313 | 0.58 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr12_+_25640480 | 0.57 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr2_+_23222939 | 0.57 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr6_-_57722816 | 0.57 |
ENSDART00000186163
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr3_+_22059066 | 0.57 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr13_+_14006118 | 0.57 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr5_-_43935460 | 0.56 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr15_+_44093286 | 0.56 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr3_+_40856095 | 0.56 |
ENSDART00000143207
|
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr9_+_1505206 | 0.56 |
ENSDART00000093427
ENSDART00000137230 |
pde11a
|
phosphodiesterase 11a |
| chr11_+_39969048 | 0.56 |
ENSDART00000193693
|
per3
|
period circadian clock 3 |
| chr19_+_42754882 | 0.55 |
ENSDART00000133711
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr24_-_20808283 | 0.55 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr17_+_24318753 | 0.55 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr19_+_3056450 | 0.54 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr22_-_38607504 | 0.54 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr15_-_45246911 | 0.53 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr10_-_41156348 | 0.53 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr7_+_19882066 | 0.53 |
ENSDART00000111144
|
TMEM151A
|
transmembrane protein 151A |
| chr24_+_792429 | 0.52 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr17_-_21441464 | 0.52 |
ENSDART00000031490
|
vax1
|
ventral anterior homeobox 1 |
| chr25_+_2776511 | 0.52 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr19_+_14454306 | 0.52 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr1_-_53407448 | 0.52 |
ENSDART00000160033
ENSDART00000172322 |
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr2_+_23823622 | 0.50 |
ENSDART00000099581
|
si:dkey-24c2.9
|
si:dkey-24c2.9 |
| chr15_+_32798333 | 0.50 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr12_-_13966184 | 0.50 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| chr23_+_30859086 | 0.50 |
ENSDART00000053661
|
zgc:198371
|
zgc:198371 |
| chr10_+_39476432 | 0.49 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr5_-_24712405 | 0.49 |
ENSDART00000033630
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr10_-_22057001 | 0.49 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr10_+_26118122 | 0.48 |
ENSDART00000079207
|
trim47
|
tripartite motif containing 47 |
| chr11_-_21363834 | 0.48 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr11_-_23322182 | 0.48 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr5_+_69950882 | 0.48 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr4_-_24019711 | 0.47 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr21_+_10021823 | 0.47 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr7_+_32021669 | 0.46 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr1_-_23557877 | 0.46 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr24_-_6345647 | 0.46 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr2_+_38147761 | 0.45 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr9_-_32158288 | 0.45 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr25_+_3994823 | 0.45 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr20_+_52458765 | 0.45 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr3_-_23596532 | 0.44 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr3_-_13068189 | 0.43 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_-_30815826 | 0.43 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr10_-_2682198 | 0.43 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr10_+_34377697 | 0.43 |
ENSDART00000189441
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr5_-_52277643 | 0.43 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr10_-_27196093 | 0.43 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr18_-_16590056 | 0.43 |
ENSDART00000143744
|
mgat4c
|
mgat4 family, member C |
| chr18_-_46881682 | 0.43 |
ENSDART00000189653
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr4_-_9764767 | 0.43 |
ENSDART00000164328
ENSDART00000147699 |
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr11_+_2687395 | 0.43 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr3_+_311833 | 0.42 |
ENSDART00000187375
|
FP326649.2
|
|
| chr8_+_2757821 | 0.42 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr19_-_27830818 | 0.42 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr13_-_44782462 | 0.42 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr22_-_20166660 | 0.42 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr20_-_47731768 | 0.41 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr21_-_18824434 | 0.41 |
ENSDART00000156333
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr18_-_20560007 | 0.41 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr5_-_41307550 | 0.40 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr11_-_2594045 | 0.40 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 1.9 | GO:0089700 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.3 | 0.9 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 2.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.8 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 1.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 1.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 0.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 1.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 1.8 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) hypothalamus cell migration(GO:0021855) |
| 0.2 | 1.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 0.6 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.2 | 1.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 1.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.9 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.3 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 1.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 2.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 0.8 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.5 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.9 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.1 | 1.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.3 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.0 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 1.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 3.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.3 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.0 | 0.6 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.6 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.3 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 3.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 1.3 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.5 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.4 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.3 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.2 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 3.5 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 5.5 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 1.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 1.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 3.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.6 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.4 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 4.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.6 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.2 | 1.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.6 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 1.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 0.5 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 3.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 3.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.0 | 0.3 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 1.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.6 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |