Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
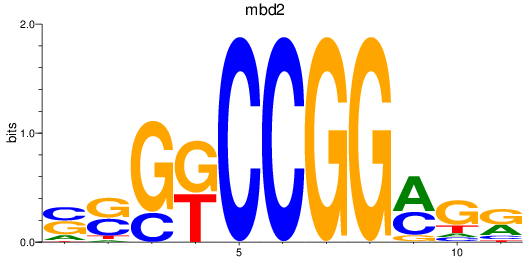
Results for mbd2
Z-value: 1.90
Transcription factors associated with mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mbd2
|
ENSDARG00000075952 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mbd2 | dr11_v1_chr5_-_1047222_1047222 | 0.29 | 6.3e-01 | Click! |
Activity profile of mbd2 motif
Sorted Z-values of mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_46763170 | 2.45 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr18_-_46464501 | 2.06 |
ENSDART00000040669
|
sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr11_-_762721 | 1.99 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr25_+_34407529 | 1.97 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr10_+_16225870 | 1.96 |
ENSDART00000164647
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr8_+_1148876 | 1.92 |
ENSDART00000148651
|
avp
|
arginine vasopressin |
| chr7_+_30867008 | 1.86 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr23_-_3681026 | 1.84 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr21_-_12272543 | 1.77 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr8_-_54223316 | 1.75 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr22_-_12862415 | 1.73 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr19_-_27966526 | 1.66 |
ENSDART00000141896
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr2_-_56635744 | 1.61 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr9_+_2452672 | 1.51 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr21_+_26697536 | 1.48 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr20_+_40457599 | 1.44 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr16_+_5678071 | 1.43 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr24_-_35699595 | 1.42 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_27966780 | 1.31 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr20_-_40717900 | 1.26 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr22_-_3564563 | 1.22 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr20_+_16743056 | 1.20 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr24_-_35699444 | 1.17 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr20_+_54738210 | 1.17 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr6_+_39222598 | 1.15 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_+_5480327 | 1.12 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr16_-_45058919 | 1.10 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr23_+_45584223 | 1.09 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr14_-_3031810 | 1.05 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr25_+_21833287 | 1.02 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr4_-_24019711 | 1.02 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr12_-_3756405 | 0.98 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr21_+_28728030 | 0.96 |
ENSDART00000097307
|
puraa
|
purine-rich element binding protein Aa |
| chr4_+_11439511 | 0.95 |
ENSDART00000150485
|
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr14_-_3032016 | 0.94 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr6_-_1874664 | 0.92 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr19_-_42045372 | 0.92 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr13_-_40120252 | 0.89 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr1_+_32521469 | 0.88 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr10_+_16225553 | 0.79 |
ENSDART00000129844
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr20_+_18943406 | 0.77 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr16_-_27161410 | 0.76 |
ENSDART00000177503
|
LO017771.1
|
|
| chr14_+_44860335 | 0.76 |
ENSDART00000091620
ENSDART00000173043 ENSDART00000091625 |
atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr5_+_32009956 | 0.75 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr2_-_32738535 | 0.75 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr17_-_26604549 | 0.72 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr4_+_16885854 | 0.72 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr16_-_42238120 | 0.69 |
ENSDART00000084730
|
zgc:162160
|
zgc:162160 |
| chr2_-_689047 | 0.68 |
ENSDART00000122732
|
foxc1a
|
forkhead box C1a |
| chr5_-_12560569 | 0.68 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr3_-_25813426 | 0.67 |
ENSDART00000039482
|
ntn1b
|
netrin 1b |
| chr9_+_24159725 | 0.67 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr17_+_53250802 | 0.67 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr13_-_1349922 | 0.66 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr15_+_36966369 | 0.66 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr2_+_23823622 | 0.65 |
ENSDART00000099581
|
si:dkey-24c2.9
|
si:dkey-24c2.9 |
| chr6_+_51932563 | 0.65 |
ENSDART00000181778
|
angpt4
|
angiopoietin 4 |
| chr12_+_25600685 | 0.65 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr5_+_71999996 | 0.65 |
ENSDART00000179933
ENSDART00000187070 |
PLPP7 (1 of many)
|
phospholipid phosphatase 7 (inactive) |
| chr2_+_50062165 | 0.63 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr18_-_51015718 | 0.63 |
ENSDART00000190698
|
LO018598.1
|
|
| chr22_+_110158 | 0.61 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr13_-_44782462 | 0.61 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr24_-_21404367 | 0.59 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr25_+_34407740 | 0.59 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr22_-_2929127 | 0.57 |
ENSDART00000123730
ENSDART00000046678 |
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr15_-_3078600 | 0.55 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr19_+_56351 | 0.54 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr8_-_53926228 | 0.54 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr14_-_24251057 | 0.52 |
ENSDART00000114169
|
bnip1a
|
BCL2 interacting protein 1a |
| chr5_+_393738 | 0.52 |
ENSDART00000161456
|
june
|
JunE proto-oncogene, AP-1 transcription factor subunit |
| chr1_-_65017 | 0.50 |
ENSDART00000168609
|
si:zfos-1011f11.2
|
si:zfos-1011f11.2 |
| chr3_-_17716322 | 0.49 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr18_+_38749547 | 0.49 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr19_-_25427255 | 0.49 |
ENSDART00000036854
|
glcci1a
|
glucocorticoid induced 1a |
| chr20_-_45062514 | 0.48 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr17_-_50010121 | 0.48 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr5_+_457729 | 0.46 |
ENSDART00000025183
|
ift74
|
intraflagellar transport 74 |
| chr7_+_53754653 | 0.44 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr2_+_35854242 | 0.44 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr22_+_35275468 | 0.43 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr14_-_30983011 | 0.41 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr9_-_5046315 | 0.40 |
ENSDART00000179087
ENSDART00000109954 |
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr14_-_45967712 | 0.40 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr1_-_64829 | 0.39 |
ENSDART00000180269
|
si:zfos-1011f11.2
|
si:zfos-1011f11.2 |
| chr22_+_35275206 | 0.39 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr8_+_54013199 | 0.37 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr22_-_36829006 | 0.37 |
ENSDART00000170256
ENSDART00000086064 |
map1sa
|
microtubule-associated protein 1Sa |
| chr18_-_127873 | 0.35 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr20_-_22193190 | 0.34 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr16_+_27564270 | 0.33 |
ENSDART00000140460
|
tmem67
|
transmembrane protein 67 |
| chr13_-_27620815 | 0.33 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr6_-_31336317 | 0.33 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_+_53755054 | 0.33 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr1_-_22803147 | 0.31 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr21_+_22833905 | 0.30 |
ENSDART00000111150
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr15_+_11693624 | 0.30 |
ENSDART00000193630
ENSDART00000161930 |
strn4
|
striatin, calmodulin binding protein 4 |
| chr9_-_21936841 | 0.30 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr23_+_39331216 | 0.30 |
ENSDART00000160957
|
kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr21_-_5393125 | 0.28 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr18_+_3579829 | 0.28 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr14_+_14992909 | 0.28 |
ENSDART00000182203
|
BX119314.1
|
|
| chr2_-_57891504 | 0.27 |
ENSDART00000139948
|
zgc:92789
|
zgc:92789 |
| chr8_+_25900049 | 0.26 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr8_+_2878756 | 0.25 |
ENSDART00000168107
|
crb2b
|
crumbs family member 2b |
| chr1_-_30689004 | 0.22 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr12_-_3773869 | 0.22 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr5_-_34616599 | 0.21 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr17_-_52636937 | 0.20 |
ENSDART00000084268
|
fam98b
|
family with sequence similarity 98, member B |
| chr7_-_9556354 | 0.18 |
ENSDART00000127974
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr12_+_763051 | 0.17 |
ENSDART00000152579
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr13_+_4888351 | 0.16 |
ENSDART00000145940
|
micu1
|
mitochondrial calcium uptake 1 |
| chr12_-_11294979 | 0.13 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr12_-_287633 | 0.12 |
ENSDART00000113335
|
map2k4b
|
mitogen-activated protein kinase kinase 4b |
| chr17_-_8673278 | 0.10 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr23_-_17450746 | 0.09 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr4_+_27018663 | 0.09 |
ENSDART00000180778
|
CR749777.1
|
|
| chr19_+_14352332 | 0.08 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr11_-_575786 | 0.08 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr19_-_45534392 | 0.08 |
ENSDART00000163920
|
trps1
|
trichorhinophalangeal syndrome I |
| chr10_-_26999482 | 0.08 |
ENSDART00000146085
|
si:dkey-88p24.11
|
si:dkey-88p24.11 |
| chr12_-_11457625 | 0.08 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr14_+_556501 | 0.08 |
ENSDART00000181289
|
nudt6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr11_-_27738489 | 0.07 |
ENSDART00000181612
|
fam120a
|
family with sequence similarity 120A |
| chr18_+_54354 | 0.07 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr13_-_51922290 | 0.06 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr10_+_16036246 | 0.06 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr6_-_19664848 | 0.05 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr9_+_53725294 | 0.05 |
ENSDART00000165991
|
cnmd
|
chondromodulin |
| chr10_-_28193642 | 0.05 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr15_+_404891 | 0.03 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr9_+_711822 | 0.03 |
ENSDART00000136627
|
ybey
|
ybeY metallopeptidase |
| chr23_-_7052362 | 0.03 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr15_-_3940671 | 0.02 |
ENSDART00000189741
|
mindy4b
|
MINDY family member 4B |
| chr9_+_44034790 | 0.01 |
ENSDART00000166110
ENSDART00000176954 |
itga4
|
integrin alpha 4 |
| chr9_+_1365747 | 0.01 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr9_+_31222026 | 0.01 |
ENSDART00000145573
|
clybl
|
citrate lyase beta like |
Network of associatons between targets according to the STRING database.
First level regulatory network of mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.6 | 1.8 | GO:0001692 | histamine metabolic process(GO:0001692) negative regulation of anion transport(GO:1903792) |
| 0.4 | 1.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.3 | 0.8 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 2.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 0.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 1.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 1.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 2.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 0.6 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.2 | 2.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.7 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.0 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.0 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.8 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.3 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 1.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.6 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 0.9 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.1 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0033338 | medial fin development(GO:0033338) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 2.0 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 2.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 3.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 2.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 1.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 2.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 1.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 2.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 1.0 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |