Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
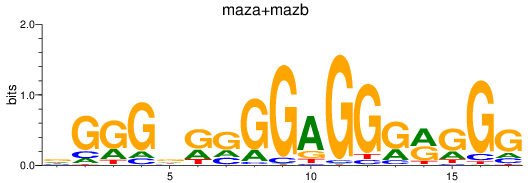
Results for maza+mazb
Z-value: 6.23
Transcription factors associated with maza+mazb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mazb
|
ENSDARG00000063555 | si_ch211-166g5.4 |
|
maza
|
ENSDARG00000087330 | MYC-associated zinc finger protein a (purine-binding transcription factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-166g5.4 | dr11_v1_chr12_-_3773869_3773869 | 0.89 | 4.5e-02 | Click! |
| maza | dr11_v1_chr3_-_21106093_21106093 | 0.76 | 1.4e-01 | Click! |
Activity profile of maza+mazb motif
Sorted Z-values of maza+mazb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_30642819 | 9.44 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr2_-_44183451 | 8.84 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr2_-_44183613 | 8.69 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr1_-_39943596 | 7.14 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr21_+_5169154 | 6.36 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr23_-_31693309 | 6.22 |
ENSDART00000146327
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr16_-_17207754 | 5.66 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr25_+_37366698 | 5.59 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr11_-_1291012 | 5.53 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr24_-_4973765 | 5.29 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr3_-_30861177 | 5.13 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr14_+_30730749 | 5.10 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr3_+_32425202 | 5.05 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr2_+_22694382 | 5.04 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr2_-_9646857 | 4.96 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr7_-_58098814 | 4.89 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr1_-_38709551 | 4.87 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr11_-_97817 | 4.85 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr3_+_54168007 | 4.82 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr17_-_46457622 | 4.76 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr21_+_26697536 | 4.75 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr11_-_44030962 | 4.74 |
ENSDART00000171910
|
FP016005.1
|
|
| chr19_-_26863626 | 4.53 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr10_-_37337579 | 4.53 |
ENSDART00000147705
|
omga
|
oligodendrocyte myelin glycoprotein a |
| chr24_+_41931585 | 4.52 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr8_+_22931427 | 4.44 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr17_+_26569601 | 4.39 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr24_+_37080771 | 4.38 |
ENSDART00000159942
|
kcnc3b
|
potassium voltage-gated channel, Shaw-related subfamily, member 3b |
| chr19_-_5254699 | 4.31 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr14_+_22172047 | 4.24 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr3_-_30625219 | 4.24 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr2_-_38125657 | 4.21 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr6_-_20875111 | 4.20 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr6_+_38381957 | 4.20 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr19_+_9533008 | 4.19 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr5_-_67878064 | 4.18 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr14_+_44545092 | 4.08 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr14_+_34486629 | 4.06 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr18_-_46763170 | 4.02 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr13_+_38430466 | 4.01 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr18_-_44610992 | 3.98 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr21_-_21373242 | 3.93 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr19_-_1961024 | 3.92 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr16_-_44349845 | 3.91 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr19_-_25301711 | 3.91 |
ENSDART00000175739
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr3_+_37827373 | 3.85 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr18_-_51015718 | 3.84 |
ENSDART00000190698
|
LO018598.1
|
|
| chr7_+_528593 | 3.81 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr1_-_31505144 | 3.76 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr16_-_27640995 | 3.68 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr8_-_14052349 | 3.66 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr1_+_44491077 | 3.60 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr4_+_20255160 | 3.59 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr3_-_19133003 | 3.57 |
ENSDART00000145215
|
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr13_-_21739142 | 3.56 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr1_-_45647846 | 3.45 |
ENSDART00000186881
|
BX511120.1
|
|
| chr13_-_638485 | 3.40 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr17_-_44247707 | 3.38 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr3_-_22212764 | 3.35 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr25_+_35706493 | 3.35 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr10_+_1849874 | 3.33 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr11_-_32723851 | 3.29 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr22_+_32120156 | 3.22 |
ENSDART00000149666
|
dock3
|
dedicator of cytokinesis 3 |
| chr3_-_46811611 | 3.20 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_-_13886952 | 3.19 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr22_+_16418688 | 3.08 |
ENSDART00000009360
|
ankrd29
|
ankyrin repeat domain 29 |
| chr11_-_44543082 | 3.06 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr25_+_19954576 | 3.01 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr13_-_32995324 | 3.01 |
ENSDART00000140542
ENSDART00000037740 |
kcnf1b
|
potassium voltage-gated channel, subfamily F, member 1b |
| chr16_-_12914288 | 2.99 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr8_-_7391721 | 2.99 |
ENSDART00000149836
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr2_+_3595333 | 2.96 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr21_-_43015383 | 2.96 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr8_+_13849999 | 2.91 |
ENSDART00000143784
|
doc2d
|
double C2-like domains, delta |
| chr3_-_36115339 | 2.89 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr6_+_55365632 | 2.88 |
ENSDART00000169240
|
pltp
|
phospholipid transfer protein |
| chr13_+_27073901 | 2.87 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr13_+_11436130 | 2.86 |
ENSDART00000169895
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_-_22660678 | 2.86 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr11_+_37137196 | 2.83 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr10_-_35542071 | 2.82 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr2_-_5014939 | 2.80 |
ENSDART00000164506
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr13_+_11440389 | 2.78 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr18_-_2727764 | 2.76 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr4_+_2619132 | 2.73 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr8_+_28900689 | 2.72 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr19_-_27777228 | 2.72 |
ENSDART00000046166
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr16_-_9383629 | 2.72 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr2_-_32555625 | 2.70 |
ENSDART00000056641
ENSDART00000137531 |
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr12_-_10705916 | 2.70 |
ENSDART00000164038
|
FO704786.1
|
|
| chr6_+_22597362 | 2.70 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr8_+_28527776 | 2.68 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr15_-_12113045 | 2.67 |
ENSDART00000159879
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr14_-_1990290 | 2.67 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr12_+_15363463 | 2.65 |
ENSDART00000133533
|
plxdc1
|
plexin domain containing 1 |
| chr7_+_427503 | 2.63 |
ENSDART00000185942
|
NRXN2 (1 of many)
|
neurexin 2 |
| chr19_+_16032383 | 2.63 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr16_-_44399335 | 2.62 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr6_-_11780070 | 2.62 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr20_+_20672163 | 2.62 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr15_-_33925851 | 2.61 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr4_+_19534833 | 2.58 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr21_-_26918901 | 2.57 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr13_+_1100197 | 2.54 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr21_-_15200556 | 2.54 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr12_+_32729470 | 2.53 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr3_-_21061931 | 2.52 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr10_-_31220558 | 2.52 |
ENSDART00000134866
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr22_+_413349 | 2.51 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr1_-_22861348 | 2.50 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr6_+_6850821 | 2.50 |
ENSDART00000166607
ENSDART00000111024 |
zgc:171558
|
zgc:171558 |
| chr17_-_14671098 | 2.49 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr22_+_18951096 | 2.48 |
ENSDART00000187010
|
hcn2b
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2b |
| chr8_+_32516160 | 2.48 |
ENSDART00000061786
|
ncs1b
|
neuronal calcium sensor 1b |
| chr3_+_37824268 | 2.46 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr6_+_54358529 | 2.46 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr3_-_28120092 | 2.45 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr13_-_33007781 | 2.43 |
ENSDART00000183671
ENSDART00000179859 |
rbm25a
|
RNA binding motif protein 25a |
| chr1_+_10221568 | 2.43 |
ENSDART00000152424
|
npy2rl
|
neuropeptide Y receptor Y2, like |
| chr10_-_7974155 | 2.42 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr19_+_2279051 | 2.42 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr16_-_8927425 | 2.40 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr23_+_6232895 | 2.39 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr4_+_20263097 | 2.37 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr25_-_19395476 | 2.37 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr7_+_27977065 | 2.35 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr10_+_31244619 | 2.34 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr17_-_43287290 | 2.34 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr5_+_38263240 | 2.33 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_-_27442339 | 2.33 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr18_-_46464501 | 2.33 |
ENSDART00000040669
|
sphkap
|
SPHK1 interactor, AKAP domain containing |
| chr7_-_24236364 | 2.32 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr17_-_26926577 | 2.31 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr3_-_21037840 | 2.31 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr24_-_20599781 | 2.31 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr24_+_27023616 | 2.30 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr22_+_12790837 | 2.28 |
ENSDART00000063055
|
mstna
|
myostatin a |
| chr16_-_16590780 | 2.27 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr7_-_69521481 | 2.26 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr15_-_5467477 | 2.25 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr25_-_11088839 | 2.25 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr10_-_26179805 | 2.24 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr12_-_31009315 | 2.24 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr14_-_2369849 | 2.24 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr13_-_31435137 | 2.23 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr19_+_30633453 | 2.22 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr6_-_35779348 | 2.22 |
ENSDART00000191159
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr7_+_19552381 | 2.21 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr18_+_21122818 | 2.20 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr5_-_21524251 | 2.18 |
ENSDART00000176793
ENSDART00000040184 |
tenm1
|
teneurin transmembrane protein 1 |
| chr16_-_34212912 | 2.17 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr24_-_31942656 | 2.16 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr9_-_54840124 | 2.16 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr23_+_39281229 | 2.15 |
ENSDART00000180120
ENSDART00000193321 |
kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr9_+_17787864 | 2.14 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr4_-_8060962 | 2.14 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr17_+_52822422 | 2.13 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr18_-_42333428 | 2.12 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr16_-_36798783 | 2.10 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr10_+_34394454 | 2.10 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr22_+_38159823 | 2.09 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr17_+_25955003 | 2.09 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr6_-_4228640 | 2.08 |
ENSDART00000162497
ENSDART00000179923 |
trak2
|
trafficking protein, kinesin binding 2 |
| chr16_-_13109749 | 2.07 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr6_-_35738836 | 2.06 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr4_-_17055950 | 2.05 |
ENSDART00000162945
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr21_-_37889727 | 2.03 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr7_-_41468942 | 2.03 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr4_+_12612145 | 2.02 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr18_+_12058403 | 2.02 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr10_-_641609 | 2.01 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr2_+_9552456 | 2.01 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr2_+_28889936 | 2.01 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr15_+_7992906 | 2.00 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr6_+_39836474 | 1.99 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr8_+_48603398 | 1.97 |
ENSDART00000074900
|
zgc:195023
|
zgc:195023 |
| chr25_-_12788370 | 1.96 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr9_+_13714379 | 1.94 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr9_-_43538328 | 1.94 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr10_+_10210455 | 1.91 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr19_-_8600383 | 1.91 |
ENSDART00000081546
|
trim46b
|
tripartite motif containing 46b |
| chr18_-_39583601 | 1.90 |
ENSDART00000125116
|
tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr23_+_23183449 | 1.88 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr18_-_19350792 | 1.86 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr4_-_1360495 | 1.85 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr7_+_46261199 | 1.85 |
ENSDART00000170390
ENSDART00000183227 |
znf536
|
zinc finger protein 536 |
| chr13_+_28854438 | 1.84 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr8_+_46418996 | 1.84 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr6_+_2951645 | 1.84 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr17_+_2063693 | 1.83 |
ENSDART00000182349
|
zgc:162989
|
zgc:162989 |
| chr8_+_25913787 | 1.82 |
ENSDART00000190257
ENSDART00000062515 |
sema3h
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3H |
| chr1_-_29045426 | 1.82 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr11_+_23933016 | 1.82 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr19_+_13375838 | 1.81 |
ENSDART00000163093
|
lrp12
|
low density lipoprotein receptor-related protein 12 |
| chr3_-_28250722 | 1.81 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr25_+_22730490 | 1.80 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr19_-_27776649 | 1.80 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr4_-_17055782 | 1.78 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| chr3_-_28075756 | 1.76 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr25_-_29134654 | 1.76 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of maza+mazb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 1.3 | 4.0 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 1.3 | 4.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 1.1 | 5.7 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.0 | 3.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 1.0 | 4.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.0 | 2.9 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.9 | 4.3 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.9 | 5.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.8 | 3.3 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.8 | 18.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.8 | 4.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.8 | 3.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.8 | 2.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.8 | 5.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.7 | 2.2 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.7 | 5.6 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.7 | 6.0 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.6 | 1.9 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.6 | 4.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.6 | 1.8 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 2.3 | GO:0060300 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.6 | 2.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.5 | 3.2 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.5 | 2.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.5 | 2.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 4.5 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.5 | 1.4 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.4 | 1.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.4 | 4.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.4 | 2.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 5.4 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.4 | 4.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.4 | 6.3 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.4 | 4.1 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.4 | 1.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.4 | 1.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 2.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.3 | 1.7 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.3 | 7.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 2.0 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.3 | 2.7 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.3 | 1.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 1.6 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.3 | 2.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.3 | 0.9 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 8.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 2.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 6.7 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.3 | 1.3 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 3.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 1.5 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 2.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 5.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 5.8 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.2 | 2.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 1.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 3.6 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.2 | 3.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 3.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 16.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 0.8 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.2 | 2.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 9.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.2 | 4.2 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.2 | 2.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.2 | 1.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 4.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 1.0 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.2 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 2.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.3 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.2 | 4.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 3.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 0.9 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 2.3 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 2.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 1.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 1.6 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 2.3 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 1.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.7 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) regulation of oligodendrocyte differentiation(GO:0048713) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 2.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 1.4 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.4 | GO:0071169 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.7 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 4.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 6.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 6.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 2.9 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 3.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 1.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 18.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.8 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 10.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 3.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.4 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.9 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.1 | 6.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 1.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 3.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.1 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 0.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.0 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.1 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.3 | GO:0007613 | memory(GO:0007613) |
| 0.1 | 2.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 1.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 1.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.2 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0060832 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) oocyte animal/vegetal axis specification(GO:0060832) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.5 | GO:0021694 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 3.6 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.6 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 1.4 | GO:0051588 | regulation of neurotransmitter secretion(GO:0046928) regulation of neurotransmitter transport(GO:0051588) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 10.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.1 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 3.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 1.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 6.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.0 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.3 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.1 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 6.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0035992 | tendon formation(GO:0035992) |
| 0.0 | 5.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.0 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.9 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 2.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.9 | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal(GO:1903322) |
| 0.0 | 0.5 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 1.9 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.8 | GO:1903522 | regulation of blood circulation(GO:1903522) |
| 0.0 | 3.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.2 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.1 | GO:0030239 | myofibril assembly(GO:0030239) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 3.7 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 2.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 15.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.5 | 2.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.5 | 3.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 7.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 4.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 18.4 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.3 | 1.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 4.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 4.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 4.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 4.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 4.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 7.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 4.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 4.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 3.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 5.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 1.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 5.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 8.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 4.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 2.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 6.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 3.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 3.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 7.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 7.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 5.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 6.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 2.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 9.8 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 11.7 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 3.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 30.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 7.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 22.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 4.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 56.6 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.7 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.6 | 9.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 6.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.5 | 4.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 1.5 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.5 | 1.4 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.4 | 1.3 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.4 | 12.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 1.2 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 6.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.4 | 2.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.8 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 1.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.3 | 2.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 9.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 16.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 7.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.3 | 6.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 1.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 2.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 8.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 4.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 2.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 3.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 10.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 4.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 6.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 7.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 8.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 9.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 4.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 4.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 4.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.5 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 2.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 0.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 1.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 6.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.4 | GO:0016934 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 4.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 6.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 3.7 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 2.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 2.9 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.1 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 4.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.3 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 2.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 3.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.9 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 3.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 6.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 5.7 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 7.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 4.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.9 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 3.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 5.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 19.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 3.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 7.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 9.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 2.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 9.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 4.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 5.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.1 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 26.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.5 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 2.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 5.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 2.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 2.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 5.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 2.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.6 | 8.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 8.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 11.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 2.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 4.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 3.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 5.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 1.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 4.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 4.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 4.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 2.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |