Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
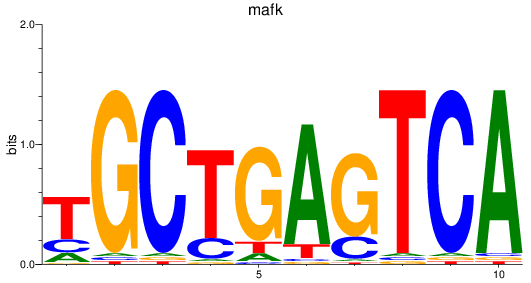
Results for mafk
Z-value: 1.62
Transcription factors associated with mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafk
|
ENSDARG00000100947 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafk | dr11_v1_chr3_-_42967698_42967698 | 0.64 | 2.5e-01 | Click! |
Activity profile of mafk motif
Sorted Z-values of mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_2020044 | 1.25 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr13_+_35746440 | 1.23 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr16_-_16182319 | 1.18 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr19_+_31771270 | 1.07 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr3_-_32818607 | 0.99 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr10_+_4235998 | 0.91 |
ENSDART00000169328
|
plppr1
|
phospholipid phosphatase related 1 |
| chr12_-_4683325 | 0.90 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr22_-_13857729 | 0.90 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr23_-_15878879 | 0.89 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr15_+_37559570 | 0.88 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr17_-_12389259 | 0.87 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr2_+_9822319 | 0.82 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr7_+_39386982 | 0.81 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr2_+_9821757 | 0.81 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr16_-_43026273 | 0.78 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr20_-_39271844 | 0.73 |
ENSDART00000192708
|
clu
|
clusterin |
| chr20_-_31905968 | 0.71 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr24_+_41940299 | 0.70 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr6_-_47246948 | 0.70 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr13_-_226109 | 0.68 |
ENSDART00000161705
ENSDART00000172744 ENSDART00000163902 ENSDART00000158208 |
rtn4b
|
reticulon 4b |
| chr19_-_6385594 | 0.68 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr16_+_14033121 | 0.68 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr24_+_24923166 | 0.67 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr4_-_5291256 | 0.67 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr11_-_10770053 | 0.66 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr14_-_17588345 | 0.65 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr20_-_47348116 | 0.65 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr20_-_47347962 | 0.65 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr23_-_27571667 | 0.64 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr10_+_466926 | 0.63 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr22_-_26595027 | 0.63 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr8_+_24861264 | 0.63 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr13_-_226393 | 0.62 |
ENSDART00000172677
|
rtn4b
|
reticulon 4b |
| chr2_+_9552456 | 0.62 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr5_+_42912966 | 0.60 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr5_-_71460556 | 0.60 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr23_-_19500559 | 0.58 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr23_-_35756115 | 0.58 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr5_+_19314574 | 0.58 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr24_-_29868151 | 0.57 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr1_-_40227166 | 0.57 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr13_-_11644806 | 0.56 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr18_+_1703984 | 0.56 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr25_+_16945348 | 0.55 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr14_+_34780886 | 0.55 |
ENSDART00000130469
ENSDART00000188806 ENSDART00000172924 ENSDART00000173266 ENSDART00000166377 ENSDART00000173371 |
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr18_-_46763170 | 0.55 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr6_+_12853655 | 0.54 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr24_-_40731305 | 0.53 |
ENSDART00000172073
|
CU633479.6
|
|
| chr8_-_22660678 | 0.53 |
ENSDART00000181258
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr1_-_1894722 | 0.51 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr3_+_7617353 | 0.51 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr6_+_40723554 | 0.51 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr3_-_21280373 | 0.50 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr23_-_29376859 | 0.49 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr16_-_27640995 | 0.49 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr10_+_16225117 | 0.49 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr4_-_9728730 | 0.48 |
ENSDART00000150265
|
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr19_-_32641725 | 0.48 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr6_+_41463786 | 0.47 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr1_-_53750522 | 0.47 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr1_+_41690402 | 0.47 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr21_-_23110841 | 0.46 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr9_-_12034444 | 0.46 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr8_-_17516448 | 0.46 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr23_+_23183449 | 0.45 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr4_+_76926059 | 0.44 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr2_-_31800521 | 0.44 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr5_-_37935607 | 0.43 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr18_+_45571378 | 0.43 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr5_+_66355153 | 0.42 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr20_-_39103119 | 0.42 |
ENSDART00000143379
|
rcan2
|
regulator of calcineurin 2 |
| chr16_-_43025885 | 0.42 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr25_-_16782394 | 0.42 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr7_+_63325819 | 0.41 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr1_+_49266886 | 0.41 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr1_-_46989650 | 0.40 |
ENSDART00000129072
ENSDART00000150507 |
si:dkey-22n8.3
|
si:dkey-22n8.3 |
| chr16_+_9779680 | 0.40 |
ENSDART00000154253
|
zgc:92912
|
zgc:92912 |
| chr5_+_36614196 | 0.39 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr2_+_25278107 | 0.39 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_-_50917255 | 0.39 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr11_+_26609110 | 0.38 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr16_-_48673938 | 0.38 |
ENSDART00000156969
|
notchl
|
notch homolog, like |
| chr17_+_22311413 | 0.38 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr19_-_6873107 | 0.38 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr7_-_52842605 | 0.37 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr4_+_6572364 | 0.37 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr6_-_35446110 | 0.36 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr11_-_9948487 | 0.36 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr13_-_23007813 | 0.36 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr14_-_30932373 | 0.36 |
ENSDART00000172988
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr6_-_12135741 | 0.36 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr18_-_5692292 | 0.35 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr9_+_28688574 | 0.35 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr5_+_28398449 | 0.35 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr1_-_25966068 | 0.35 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr9_+_34380299 | 0.35 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr23_-_10745288 | 0.35 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr14_-_30490763 | 0.35 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr8_+_36948256 | 0.35 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr5_-_21030934 | 0.35 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr16_-_9869056 | 0.35 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr19_+_30990129 | 0.35 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr5_-_63109232 | 0.35 |
ENSDART00000115128
|
usp2b
|
ubiquitin specific peptidase 2b |
| chr23_+_13814978 | 0.35 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr10_+_29963518 | 0.34 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr7_+_62004048 | 0.34 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr21_-_31143903 | 0.34 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr13_+_24519175 | 0.34 |
ENSDART00000190790
|
sertad2b
|
SERTA domain containing 2b |
| chr1_+_51039558 | 0.33 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr22_-_4760187 | 0.33 |
ENSDART00000081969
|
rad23ab
|
RAD23 homolog A, nucleotide excision repair protein b |
| chr7_+_17992455 | 0.33 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr14_+_24840669 | 0.33 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr24_+_26039464 | 0.32 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr3_-_21061931 | 0.32 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr25_+_6122823 | 0.32 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr16_+_14029283 | 0.32 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr5_-_62940851 | 0.31 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr19_-_10243148 | 0.31 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr24_-_21587335 | 0.31 |
ENSDART00000091528
|
gpr12
|
G protein-coupled receptor 12 |
| chr19_-_17658160 | 0.31 |
ENSDART00000151766
ENSDART00000170790 ENSDART00000186678 ENSDART00000188045 ENSDART00000176980 ENSDART00000166313 ENSDART00000188589 |
thrb
|
thyroid hormone receptor beta |
| chr5_-_19052184 | 0.31 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr7_-_52842007 | 0.30 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr19_+_1510971 | 0.30 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr23_-_9965148 | 0.30 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr16_-_13613475 | 0.30 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr14_-_48588422 | 0.30 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr13_-_40499296 | 0.30 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr23_+_23182037 | 0.30 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr7_+_20298333 | 0.30 |
ENSDART00000023089
ENSDART00000131019 |
acadvl
|
acyl-CoA dehydrogenase very long chain |
| chr18_-_14937211 | 0.30 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr21_-_11820379 | 0.30 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr7_+_69841017 | 0.29 |
ENSDART00000169107
|
FO818704.1
|
|
| chr15_+_17100412 | 0.29 |
ENSDART00000154418
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr19_+_37701450 | 0.29 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr25_-_16818978 | 0.29 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr19_+_31404686 | 0.29 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr17_-_1407593 | 0.29 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr18_-_7456378 | 0.29 |
ENSDART00000081459
|
pdp2
|
putative pyruvate dehydrogenase phosphatase isoenzyme 2 |
| chr13_+_30506781 | 0.28 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr15_-_16679517 | 0.28 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr1_-_25966411 | 0.28 |
ENSDART00000193375
|
synpo2b
|
synaptopodin 2b |
| chr22_-_31059670 | 0.28 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr6_-_14292307 | 0.28 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr24_+_12983434 | 0.28 |
ENSDART00000145214
ENSDART00000146911 ENSDART00000066700 |
eloca
|
elongin C paralog a |
| chr14_-_30490465 | 0.28 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr22_+_28446557 | 0.28 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr5_-_26493253 | 0.28 |
ENSDART00000088222
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr16_-_17162485 | 0.28 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr10_+_15608326 | 0.27 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr12_+_48216662 | 0.27 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr21_+_9576176 | 0.27 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr3_+_26064091 | 0.27 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr18_+_31410652 | 0.27 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr5_+_21931124 | 0.26 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr16_-_17162843 | 0.26 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr18_+_34478959 | 0.26 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr23_+_29966466 | 0.26 |
ENSDART00000143583
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr5_-_31928913 | 0.26 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr8_-_34051548 | 0.26 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr10_-_7974155 | 0.26 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr7_-_40630698 | 0.26 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr21_-_13123176 | 0.26 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr7_+_756942 | 0.26 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr24_+_8904741 | 0.26 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr18_-_19405616 | 0.26 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr22_+_28446365 | 0.25 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr12_-_19153177 | 0.25 |
ENSDART00000057125
|
tefa
|
thyrotrophic embryonic factor a |
| chr14_-_34771371 | 0.25 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr3_+_32391540 | 0.25 |
ENSDART00000156608
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr6_-_6254432 | 0.25 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr17_-_45247151 | 0.25 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr8_-_34052019 | 0.25 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr8_-_13211527 | 0.24 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr10_+_2684958 | 0.24 |
ENSDART00000112019
|
setd9
|
SET domain containing 9 |
| chr12_-_19862912 | 0.24 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr6_+_21395051 | 0.24 |
ENSDART00000017774
|
cacng5a
|
calcium channel, voltage-dependent, gamma subunit 5a |
| chr15_-_39820491 | 0.24 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr2_-_24962002 | 0.23 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr20_-_19422496 | 0.23 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr22_-_12161095 | 0.23 |
ENSDART00000158398
|
tmem163b
|
transmembrane protein 163b |
| chr25_+_10485103 | 0.22 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr17_-_11439815 | 0.22 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr9_+_33261330 | 0.22 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr2_-_16562505 | 0.22 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr6_+_41096058 | 0.21 |
ENSDART00000028373
|
fkbp5
|
FK506 binding protein 5 |
| chr10_+_17235370 | 0.21 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr17_-_12966907 | 0.21 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr11_+_13528437 | 0.21 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr7_+_17992640 | 0.21 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr5_-_16996482 | 0.20 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr11_-_36009924 | 0.20 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr2_+_42592572 | 0.20 |
ENSDART00000179722
|
march11
|
membrane-associated ring finger (C3HC4) 11 |
| chr6_-_21534301 | 0.20 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr3_-_18189283 | 0.20 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr15_+_28318005 | 0.20 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr23_+_3731375 | 0.20 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr2_-_9544161 | 0.20 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr2_-_155270 | 0.20 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr2_-_4797512 | 0.20 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr3_-_24205339 | 0.20 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.2 | 0.6 | GO:0015824 | proline transport(GO:0015824) |
| 0.2 | 0.5 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.4 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.3 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.7 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.3 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.3 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.3 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.3 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.6 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.4 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0090248 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.0 | 0.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.3 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.5 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.2 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.6 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.3 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 1.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.7 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.2 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.6 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.3 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |