Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
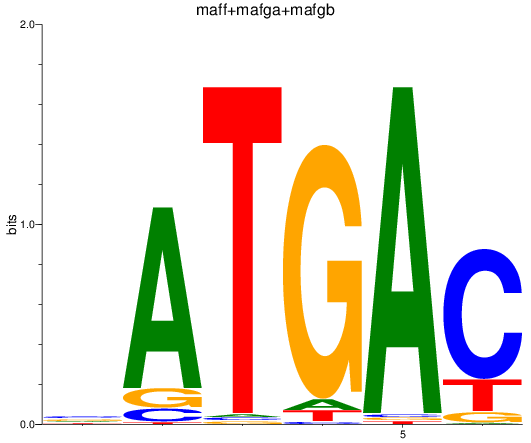
Results for maff+mafga+mafgb
Z-value: 1.38
Transcription factors associated with maff+mafga+mafgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafga
|
ENSDARG00000018109 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
|
maff
|
ENSDARG00000028957 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
|
mafgb
|
ENSDARG00000100097 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
|
maff
|
ENSDARG00000111050 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafgb | dr11_v1_chr11_+_45299447_45299447 | 0.68 | 2.1e-01 | Click! |
| mafga | dr11_v1_chr3_-_55404985_55404985 | 0.58 | 3.0e-01 | Click! |
| maff | dr11_v1_chr12_-_19346678_19346678 | 0.42 | 4.8e-01 | Click! |
Activity profile of maff+mafga+mafgb motif
Sorted Z-values of maff+mafga+mafgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_36040820 | 1.05 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr3_+_32526799 | 1.02 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_15907458 | 0.91 |
ENSDART00000163525
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr3_+_15907297 | 0.88 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr7_+_25059845 | 0.88 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr19_-_19339285 | 0.88 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr21_+_9666347 | 0.83 |
ENSDART00000163853
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr13_+_36146415 | 0.82 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr6_+_27667359 | 0.81 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr21_-_45613564 | 0.77 |
ENSDART00000160324
|
LO018363.1
|
|
| chr6_-_35446110 | 0.74 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr21_+_18353703 | 0.73 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr23_-_27345425 | 0.71 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr2_+_20332044 | 0.71 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr2_-_24996441 | 0.70 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr21_-_43606502 | 0.70 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr17_+_27134806 | 0.69 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr23_-_29502287 | 0.69 |
ENSDART00000141075
ENSDART00000053807 |
kif1b
|
kinesin family member 1B |
| chr7_-_24491614 | 0.69 |
ENSDART00000131063
|
si:dkeyp-75h12.5
|
si:dkeyp-75h12.5 |
| chr10_-_27049170 | 0.68 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr15_-_44512461 | 0.68 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr16_-_12173554 | 0.66 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr11_+_25112269 | 0.66 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr23_-_4915118 | 0.65 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr16_-_29452039 | 0.65 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr6_-_12135741 | 0.64 |
ENSDART00000155090
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr19_+_25649626 | 0.63 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr14_-_30490763 | 0.62 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr19_-_1961024 | 0.61 |
ENSDART00000108784
|
mturn
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr24_+_9744012 | 0.61 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr21_+_20901505 | 0.59 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr20_+_27393668 | 0.59 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr3_-_32320537 | 0.59 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr6_-_51101834 | 0.59 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr10_+_1638876 | 0.58 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr12_-_10300101 | 0.57 |
ENSDART00000126428
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr18_+_3332999 | 0.57 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr19_-_9867001 | 0.56 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr5_+_27137473 | 0.54 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr7_+_67486807 | 0.54 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr21_-_28340977 | 0.53 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr6_-_31348999 | 0.53 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_-_878473 | 0.53 |
ENSDART00000173567
|
si:cabz01080528.1
|
si:cabz01080528.1 |
| chr8_+_53452681 | 0.52 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr16_+_10777116 | 0.52 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr1_+_57331813 | 0.51 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr8_+_14792830 | 0.51 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr22_-_11136625 | 0.51 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr5_+_65491390 | 0.51 |
ENSDART00000159921
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr10_+_29698467 | 0.50 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr7_+_23515966 | 0.48 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr20_+_20637866 | 0.47 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr23_+_216012 | 0.46 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr17_-_22062364 | 0.46 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr2_+_20331445 | 0.46 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr2_+_55916911 | 0.45 |
ENSDART00000189483
ENSDART00000183647 ENSDART00000083470 |
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr21_-_24632778 | 0.45 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr1_-_10048514 | 0.45 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr3_+_29600917 | 0.45 |
ENSDART00000048867
|
sstr3
|
somatostatin receptor 3 |
| chr14_-_2355833 | 0.45 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr8_+_53423408 | 0.45 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr4_-_13156971 | 0.44 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr17_+_12698532 | 0.44 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr18_-_8579907 | 0.44 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr2_-_32356539 | 0.43 |
ENSDART00000169316
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr2_+_36862473 | 0.43 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr2_-_24369087 | 0.43 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr13_+_40034176 | 0.42 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr24_-_32408404 | 0.42 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr24_-_39771667 | 0.42 |
ENSDART00000181867
|
GFOD1
|
si:ch211-276f18.2 |
| chr7_-_57933736 | 0.41 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr7_+_72882446 | 0.41 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr5_+_23118470 | 0.40 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr9_-_18424844 | 0.40 |
ENSDART00000154351
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr3_+_31933893 | 0.40 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr4_-_77267787 | 0.39 |
ENSDART00000190346
|
zgc:174310
|
zgc:174310 |
| chr11_-_41130239 | 0.39 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr21_+_9576176 | 0.39 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr6_-_10780698 | 0.38 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr13_+_28675686 | 0.38 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr7_-_18730474 | 0.38 |
ENSDART00000170374
|
si:dkey-38l22.2
|
si:dkey-38l22.2 |
| chr10_-_20175118 | 0.38 |
ENSDART00000014417
|
htr7b
|
5-hydroxytryptamine (serotonin) receptor 7b |
| chr20_+_20638034 | 0.37 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr3_+_25508247 | 0.37 |
ENSDART00000039448
|
mchr1b
|
melanin-concentrating hormone receptor 1b |
| chr6_+_34512313 | 0.37 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr19_-_29887629 | 0.36 |
ENSDART00000066123
|
kpna6
|
karyopherin alpha 6 (importin alpha 7) |
| chr6_-_33685325 | 0.36 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr21_+_13383413 | 0.35 |
ENSDART00000151345
|
zgc:113162
|
zgc:113162 |
| chr21_+_10794914 | 0.35 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr18_+_7286788 | 0.35 |
ENSDART00000022998
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr3_-_60142530 | 0.34 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr13_-_36545258 | 0.34 |
ENSDART00000186171
|
FP103009.1
|
|
| chr24_+_38301080 | 0.34 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr9_-_4506819 | 0.34 |
ENSDART00000113975
|
kcnj3a
|
potassium inwardly-rectifying channel, subfamily J, member 3a |
| chr2_+_23222939 | 0.34 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr13_+_28618086 | 0.34 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr4_-_64871624 | 0.34 |
ENSDART00000124817
|
znf988
|
zinc finger protein 988 |
| chr8_+_50983551 | 0.33 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr9_-_30346279 | 0.32 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr7_-_42206720 | 0.32 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr17_-_24564674 | 0.32 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr17_+_20204122 | 0.32 |
ENSDART00000078672
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr20_-_27864964 | 0.32 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr18_-_19350792 | 0.32 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr1_+_6817292 | 0.32 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr3_-_22829710 | 0.31 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr7_+_42206543 | 0.31 |
ENSDART00000112543
|
phkb
|
phosphorylase kinase, beta |
| chr19_+_8985230 | 0.30 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr7_+_34290051 | 0.30 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr5_+_36870737 | 0.29 |
ENSDART00000145182
|
slc8a2a
|
solute carrier family 8 (sodium/calcium exchanger), member 2a |
| chr3_+_24189804 | 0.29 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr6_+_52064304 | 0.29 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr5_-_18513950 | 0.29 |
ENSDART00000145878
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr10_+_21758811 | 0.29 |
ENSDART00000188827
|
pcdh1g11
|
protocadherin 1 gamma 11 |
| chr24_+_32472155 | 0.29 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr18_-_12295092 | 0.28 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr12_-_22509069 | 0.28 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr5_-_21422390 | 0.28 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr2_-_32505091 | 0.28 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr9_-_16133263 | 0.28 |
ENSDART00000077187
|
myo1b
|
myosin IB |
| chr19_+_32947910 | 0.28 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase H+ transporting V1 subunit C1b |
| chr3_+_22905341 | 0.28 |
ENSDART00000111435
|
hdac5
|
histone deacetylase 5 |
| chr8_+_52491436 | 0.27 |
ENSDART00000164298
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr10_+_42733210 | 0.27 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr5_+_25681174 | 0.27 |
ENSDART00000134773
|
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr17_-_45009782 | 0.27 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr18_+_8231138 | 0.27 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr8_+_39607466 | 0.27 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr11_-_22372072 | 0.26 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr2_+_10642047 | 0.26 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr11_-_18791834 | 0.26 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr3_+_33300522 | 0.26 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr9_+_27411502 | 0.26 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr17_-_51938663 | 0.25 |
ENSDART00000179784
|
ERG28
|
ergosterol biosynthesis 28 homolog |
| chr25_+_20077225 | 0.25 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr12_-_48960308 | 0.25 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr6_+_18367388 | 0.25 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr16_-_21047872 | 0.25 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr23_-_33640959 | 0.25 |
ENSDART00000187599
ENSDART00000189475 |
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr5_-_31856681 | 0.25 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr12_+_6041575 | 0.25 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr19_-_13286722 | 0.24 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr2_+_24700922 | 0.24 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr19_-_41991104 | 0.24 |
ENSDART00000087055
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr23_+_2760573 | 0.24 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr7_-_19600181 | 0.24 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr5_+_25680845 | 0.24 |
ENSDART00000139701
ENSDART00000009952 |
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr20_-_45040916 | 0.24 |
ENSDART00000190001
|
klhl29
|
kelch-like family member 29 |
| chr9_+_17423941 | 0.24 |
ENSDART00000112884
ENSDART00000155233 |
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr9_+_33174064 | 0.23 |
ENSDART00000131910
ENSDART00000078256 |
dopey2
|
dopey family member 2 |
| chr9_-_25255490 | 0.23 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr16_+_26777473 | 0.23 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr8_-_54304381 | 0.22 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr3_+_32403758 | 0.22 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr21_+_30194904 | 0.21 |
ENSDART00000137023
ENSDART00000078403 |
BRD8
|
si:ch211-59d17.3 |
| chr3_-_32169754 | 0.21 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr9_+_33158191 | 0.21 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr3_+_24190207 | 0.21 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr15_+_36966369 | 0.21 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr5_-_41307550 | 0.21 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr14_-_48348973 | 0.21 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr10_-_32877348 | 0.21 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr5_+_61738276 | 0.20 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr17_+_42027969 | 0.20 |
ENSDART00000147563
|
kiz
|
kizuna centrosomal protein |
| chr13_+_51579851 | 0.20 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr1_-_11291324 | 0.20 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr12_-_31422433 | 0.19 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr17_+_50657509 | 0.19 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr20_+_14968031 | 0.19 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr20_+_32224405 | 0.19 |
ENSDART00000062993
ENSDART00000147448 |
sesn1
|
sestrin 1 |
| chr1_+_35790082 | 0.19 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr1_+_52518176 | 0.19 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr15_-_5563551 | 0.19 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr14_-_48033073 | 0.19 |
ENSDART00000193115
ENSDART00000169300 ENSDART00000188036 ENSDART00000183432 ENSDART00000180973 |
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr13_-_33207367 | 0.19 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr23_+_37185247 | 0.19 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr7_+_7151832 | 0.19 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr7_-_72423666 | 0.19 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr4_+_25912308 | 0.19 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr20_+_27331008 | 0.19 |
ENSDART00000141486
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr25_+_33063762 | 0.19 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr16_-_26435431 | 0.18 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr7_+_22702437 | 0.18 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr12_+_27024676 | 0.18 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr12_+_49125510 | 0.18 |
ENSDART00000185804
|
FO704607.1
|
|
| chr4_-_41269844 | 0.18 |
ENSDART00000186177
|
CR388165.2
|
|
| chr7_+_42208859 | 0.18 |
ENSDART00000148643
|
phkb
|
phosphorylase kinase, beta |
| chr16_+_43077909 | 0.18 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr10_-_1320114 | 0.18 |
ENSDART00000073617
|
opn4xa
|
opsin 4xa |
| chr25_-_554142 | 0.18 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr6_-_36182115 | 0.18 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr4_+_25912654 | 0.18 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr13_+_10023256 | 0.18 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr17_+_24722646 | 0.17 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr14_+_34547554 | 0.17 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr10_-_27009413 | 0.17 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr22_-_24858042 | 0.17 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr13_+_1015749 | 0.17 |
ENSDART00000190982
|
prokr1b
|
prokineticin receptor 1b |
| chr21_-_2209012 | 0.17 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr10_-_26512742 | 0.17 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of maff+mafga+mafgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.7 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 0.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.6 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.4 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.4 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 1.3 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.2 | GO:1990253 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 2.0 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0009202 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.5 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.6 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 1.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.3 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.2 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |