Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
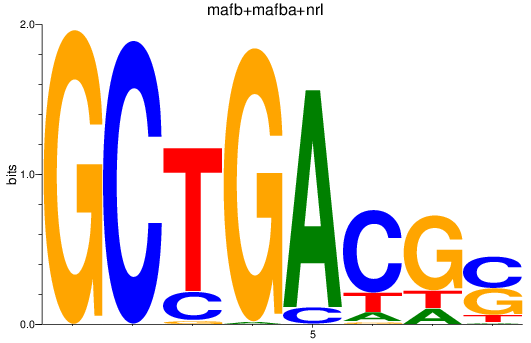
Results for mafb+mafba+nrl
Z-value: 3.51
Transcription factors associated with mafb+mafba+nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafba
|
ENSDARG00000017121 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
|
mafb
|
ENSDARG00000076520 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
|
nrl
|
ENSDARG00000100466 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nrl | dr11_v1_chr20_+_661037_661037 | 0.81 | 9.9e-02 | Click! |
| mafba | dr11_v1_chr23_-_3409140_3409140 | 0.68 | 2.1e-01 | Click! |
| mafb | dr11_v1_chr25_+_36674715_36674715 | 0.33 | 5.8e-01 | Click! |
Activity profile of mafb+mafba+nrl motif
Sorted Z-values of mafb+mafba+nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_1291012 | 4.07 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr6_-_60147517 | 3.66 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr8_-_14052349 | 3.60 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr10_-_7858553 | 3.18 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr19_+_233143 | 3.09 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr25_-_3503164 | 2.84 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr15_+_16897554 | 2.62 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr25_-_4482449 | 2.56 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr7_-_26087807 | 2.47 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr2_-_44720551 | 2.31 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr23_+_45579497 | 2.27 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr9_-_296169 | 2.18 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr19_+_37925616 | 2.18 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr21_+_41697552 | 2.14 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr8_+_24854600 | 2.12 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr1_+_36437585 | 2.07 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr1_-_21344478 | 2.05 |
ENSDART00000077805
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr19_-_5254699 | 2.03 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr14_+_49135264 | 2.03 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr7_+_72882446 | 2.00 |
ENSDART00000131113
ENSDART00000174223 |
ADGRL3
|
adhesion G protein-coupled receptor L3 |
| chr22_-_38543630 | 1.99 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr4_+_26496489 | 1.99 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr4_+_19534833 | 1.96 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr22_+_18816662 | 1.96 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr1_+_54673846 | 1.95 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr6_+_58543336 | 1.92 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr13_+_3954540 | 1.86 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr24_-_4973765 | 1.85 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr20_-_54462551 | 1.82 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr16_-_44349845 | 1.81 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr25_-_3503458 | 1.79 |
ENSDART00000173269
|
PRKAR2B
|
si:ch211-272n13.7 |
| chr2_+_6926100 | 1.79 |
ENSDART00000153289
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr6_-_24358732 | 1.78 |
ENSDART00000159595
|
ephx4
|
epoxide hydrolase 4 |
| chr8_-_23416362 | 1.77 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr19_+_23982466 | 1.76 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr20_+_88168 | 1.76 |
ENSDART00000149283
|
zgc:112001
|
zgc:112001 |
| chr5_-_12525441 | 1.76 |
ENSDART00000177490
ENSDART00000164190 |
ksr2
|
kinase suppressor of ras 2 |
| chr7_-_38612230 | 1.75 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr2_+_31957554 | 1.75 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr17_-_37214196 | 1.74 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr14_+_33458294 | 1.73 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr13_+_3954715 | 1.73 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr15_+_2559875 | 1.72 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr7_+_23875269 | 1.70 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr8_+_29267093 | 1.68 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr14_-_30642819 | 1.66 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr23_+_21067684 | 1.65 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr13_-_27038407 | 1.64 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr11_-_3494964 | 1.64 |
ENSDART00000162369
|
acap3b
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3b |
| chr5_+_36614196 | 1.63 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr25_+_19999623 | 1.61 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr6_-_13188667 | 1.60 |
ENSDART00000191654
|
adam23a
|
ADAM metallopeptidase domain 23a |
| chr3_-_30118856 | 1.59 |
ENSDART00000109953
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr1_+_49266886 | 1.58 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr23_-_26077038 | 1.58 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr21_-_23475361 | 1.58 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr17_+_9310259 | 1.57 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr8_+_53464216 | 1.57 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr13_+_14006118 | 1.56 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr24_-_31942656 | 1.56 |
ENSDART00000180308
ENSDART00000016879 |
c1ql3a
|
complement component 1, q subcomponent-like 3a |
| chr16_-_12984631 | 1.55 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr22_+_17359346 | 1.54 |
ENSDART00000145434
|
gpr52
|
G protein-coupled receptor 52 |
| chr10_+_1052591 | 1.51 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr5_-_28915130 | 1.50 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr17_+_29345606 | 1.49 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr17_-_12336987 | 1.48 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_-_17585618 | 1.48 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr1_-_59251874 | 1.47 |
ENSDART00000168919
|
olfm2b
|
olfactomedin 2b |
| chr23_-_45049044 | 1.44 |
ENSDART00000067630
|
SH3GL2 (1 of many)
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr4_+_12113105 | 1.41 |
ENSDART00000182399
|
tmem178b
|
transmembrane protein 178B |
| chr13_-_35051897 | 1.40 |
ENSDART00000129559
|
btbd3b
|
BTB (POZ) domain containing 3b |
| chr5_-_21030934 | 1.39 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr21_-_45588720 | 1.39 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr14_-_52521460 | 1.39 |
ENSDART00000172110
|
GPR151
|
G protein-coupled receptor 151 |
| chr23_-_3674443 | 1.38 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr5_-_12524996 | 1.34 |
ENSDART00000142258
|
si:ch73-263f13.1
|
si:ch73-263f13.1 |
| chr1_-_57280585 | 1.33 |
ENSDART00000152220
|
si:dkey-27j5.5
|
si:dkey-27j5.5 |
| chr13_-_11644806 | 1.32 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr4_+_8797197 | 1.32 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr5_+_30624183 | 1.32 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr12_+_32292564 | 1.32 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr23_-_29667544 | 1.31 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr7_+_58686860 | 1.31 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr9_+_49659114 | 1.29 |
ENSDART00000189643
|
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr21_+_42226113 | 1.29 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr4_-_191736 | 1.29 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr3_-_19133003 | 1.28 |
ENSDART00000145215
|
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr23_-_28141419 | 1.27 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr9_+_53707240 | 1.26 |
ENSDART00000171490
|
PCDH8
|
si:ch211-199f5.1 |
| chr2_-_9059955 | 1.26 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr16_+_34528409 | 1.26 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr21_+_26726936 | 1.25 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr8_+_50983551 | 1.25 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr10_+_35953068 | 1.25 |
ENSDART00000015279
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr12_-_11570 | 1.24 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr5_-_22501663 | 1.24 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr6_-_10776254 | 1.23 |
ENSDART00000184008
|
gpr155b
|
G protein-coupled receptor 155b |
| chr3_+_14317802 | 1.23 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr18_+_14477740 | 1.23 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr14_-_2050057 | 1.22 |
ENSDART00000112875
|
PCDHB15
|
protocadherin beta 15 |
| chr20_-_40717900 | 1.22 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr6_+_59642695 | 1.21 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr9_-_44295071 | 1.21 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr20_+_52389858 | 1.20 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr10_+_44641599 | 1.20 |
ENSDART00000172128
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr5_-_70625697 | 1.20 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr20_-_37629084 | 1.20 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr19_-_27261102 | 1.19 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr20_+_48803248 | 1.19 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr23_-_7799184 | 1.19 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr7_-_24699985 | 1.19 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr5_+_45976130 | 1.19 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr19_-_10081047 | 1.19 |
ENSDART00000128837
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr18_-_85294 | 1.18 |
ENSDART00000044387
|
gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr4_-_21851473 | 1.18 |
ENSDART00000019748
|
lin7a
|
lin-7 homolog A (C. elegans) |
| chr12_-_29624638 | 1.18 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr22_+_635813 | 1.18 |
ENSDART00000179067
|
CU856139.1
|
|
| chr17_-_12385308 | 1.18 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr2_-_24444838 | 1.18 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr24_-_29997145 | 1.17 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr20_+_54383838 | 1.17 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr12_-_45533034 | 1.17 |
ENSDART00000113004
|
CABZ01068356.1
|
|
| chr5_+_57743815 | 1.17 |
ENSDART00000005090
|
alg9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr4_-_27350820 | 1.16 |
ENSDART00000145806
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr21_-_31143903 | 1.16 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr20_-_34801181 | 1.16 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr23_-_11870962 | 1.15 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr10_+_16188761 | 1.14 |
ENSDART00000193244
|
ctxn3
|
cortexin 3 |
| chr18_-_1228688 | 1.14 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr2_-_58193381 | 1.14 |
ENSDART00000159040
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr23_-_14990865 | 1.13 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr11_+_30161168 | 1.13 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr8_-_4596662 | 1.13 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr6_-_58828398 | 1.12 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr22_-_600016 | 1.12 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr5_+_20148671 | 1.11 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr23_-_44494401 | 1.11 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr23_+_25210953 | 1.11 |
ENSDART00000132799
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr24_+_39129316 | 1.09 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_-_9882821 | 1.09 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr19_-_9712530 | 1.08 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr16_-_6424816 | 1.08 |
ENSDART00000164864
ENSDART00000141860 |
mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr10_+_39164638 | 1.08 |
ENSDART00000188997
|
CU633908.1
|
|
| chr2_+_47623202 | 1.07 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr24_-_35707552 | 1.07 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr23_+_2560005 | 1.07 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr3_-_18711288 | 1.06 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr25_-_12906872 | 1.06 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
| chr17_-_43286903 | 1.06 |
ENSDART00000176637
|
EML5
|
si:dkey-1f12.3 |
| chr14_-_2206476 | 1.06 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr19_+_32947910 | 1.05 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase H+ transporting V1 subunit C1b |
| chr8_+_1082100 | 1.05 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr1_+_7679328 | 1.05 |
ENSDART00000163488
ENSDART00000190070 |
en1b
|
engrailed homeobox 1b |
| chr12_-_3840664 | 1.05 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr4_-_8882572 | 1.04 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr8_+_53423408 | 1.04 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr6_+_41255485 | 1.04 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr9_-_41323746 | 1.04 |
ENSDART00000140564
|
glsb
|
glutaminase b |
| chr9_-_23217196 | 1.03 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr13_+_15657911 | 1.02 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr1_+_29281764 | 1.01 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr15_-_10341048 | 1.01 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr20_-_31905968 | 1.01 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr14_+_1365097 | 1.01 |
ENSDART00000083797
ENSDART00000176087 |
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr12_+_42574148 | 1.01 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr5_+_71999996 | 1.01 |
ENSDART00000179933
ENSDART00000187070 |
PLPP7 (1 of many)
|
phospholipid phosphatase 7 (inactive) |
| chr22_-_22416337 | 1.00 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr4_+_23223881 | 1.00 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr9_-_327901 | 1.00 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr12_-_26383242 | 1.00 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr8_+_25254435 | 0.99 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr1_-_20271138 | 0.99 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr15_-_19771981 | 0.99 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr1_+_29858032 | 0.99 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr19_+_33093395 | 0.99 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr1_+_59154521 | 0.98 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr1_+_38758261 | 0.98 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr2_-_56635744 | 0.98 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr15_+_46386261 | 0.98 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr3_+_20156956 | 0.98 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr20_-_20931197 | 0.97 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr18_+_22793465 | 0.97 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr8_+_54055390 | 0.97 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr17_-_7861219 | 0.97 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr5_+_36415978 | 0.96 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr15_+_24212847 | 0.96 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr7_-_38477235 | 0.96 |
ENSDART00000084355
|
zgc:165481
|
zgc:165481 |
| chr4_-_19028861 | 0.96 |
ENSDART00000166374
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr6_-_43092175 | 0.95 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr19_+_33093577 | 0.94 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr18_+_30028135 | 0.94 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr24_+_24461558 | 0.94 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr4_+_11439511 | 0.94 |
ENSDART00000150485
|
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr3_-_35801146 | 0.94 |
ENSDART00000150911
|
caskin1
|
CASK interacting protein 1 |
| chr18_+_38908903 | 0.94 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr4_+_4816695 | 0.93 |
ENSDART00000136962
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr20_-_53321499 | 0.93 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr25_+_35706493 | 0.93 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr9_+_30890549 | 0.93 |
ENSDART00000101070
|
dachd
|
dachshund d |
| chr12_-_4672708 | 0.92 |
ENSDART00000152659
ENSDART00000186076 |
si:ch211-255p10.4
|
si:ch211-255p10.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafb+mafba+nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.6 | 4.3 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.6 | 2.5 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.5 | 1.6 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.5 | 1.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.5 | 3.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 2.0 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 3.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.3 | 1.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.7 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.3 | 1.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 1.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.3 | 0.9 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 0.9 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.3 | 0.8 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.3 | 2.0 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 1.7 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.3 | 1.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.3 | 0.8 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.3 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 2.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.3 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 1.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 1.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 2.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 0.8 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.2 | 1.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 1.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 2.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 2.6 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.7 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.7 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.1 | 1.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.5 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.8 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 1.0 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 2.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.1 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.1 | 1.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 2.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.0 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.5 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.3 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 1.5 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 0.9 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.6 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.9 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.3 | GO:0035992 | tendon formation(GO:0035992) |
| 0.1 | 0.5 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 1.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.3 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.6 | GO:1903400 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 3.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 1.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.4 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 1.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 3.8 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 3.0 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 0.2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 1.0 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.3 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.6 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.9 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 1.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 3.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 1.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 2.7 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.2 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.1 | 0.6 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 1.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 0.7 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 1.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 0.4 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 1.0 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 1.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 11.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.6 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 0.4 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.2 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 1.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 4.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.3 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0048387 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.2 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 6.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 1.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.9 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.6 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 1.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.0 | 0.3 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.0 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 1.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 3.2 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.9 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.6 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0050868 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.1 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.4 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.8 | GO:0046058 | cAMP biosynthetic process(GO:0006171) cAMP metabolic process(GO:0046058) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 4.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.6 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.0 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.2 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break processing(GO:0000706) meiotic DNA double-strand break formation(GO:0042138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.4 | 1.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 1.9 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 1.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 0.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 3.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 0.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 9.9 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 1.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 1.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.4 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 3.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.3 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 2.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.5 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 2.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 6.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 1.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 3.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 4.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.1 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 2.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.7 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 5.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 4.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.2 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.0 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 8.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.5 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 3.3 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.0 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.5 | 1.4 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.4 | 2.5 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.4 | 3.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 1.8 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 1.7 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.3 | 1.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 1.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 0.9 | GO:0005333 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.3 | 3.7 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.3 | 3.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 8.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 0.8 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.2 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.5 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 8.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 1.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 2.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.7 | GO:0099583 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.7 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 2.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 3.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 2.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 2.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 2.6 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 1.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 3.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.7 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.9 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.0 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.7 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 8.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 1.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.0 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 2.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 4.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 3.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 5.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |