Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
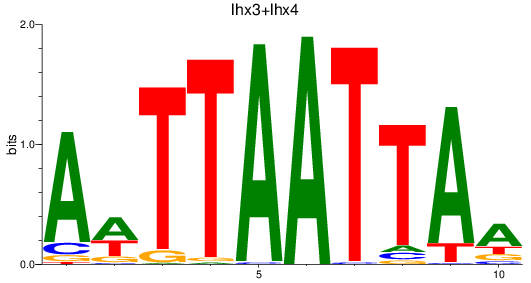
Results for lhx3+lhx4
Z-value: 1.36
Transcription factors associated with lhx3+lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx3
|
ENSDARG00000003803 | LIM homeobox 3 |
|
lhx4
|
ENSDARG00000039458 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx4 | dr11_v1_chr8_-_14484599_14484599 | 0.95 | 1.3e-02 | Click! |
| LHX3 | dr11_v1_chr5_+_71802014_71802014 | 0.84 | 7.3e-02 | Click! |
Activity profile of lhx3+lhx4 motif
Sorted Z-values of lhx3+lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_9669717 | 2.51 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr21_-_20939488 | 2.00 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr9_+_34641237 | 1.75 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr20_+_34933183 | 1.51 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr15_-_44601331 | 1.34 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr9_-_42418470 | 1.30 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr10_-_27049170 | 1.27 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr9_+_54039006 | 1.19 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr21_+_3093419 | 1.18 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr4_-_5019113 | 1.13 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr8_-_34052019 | 1.12 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr18_+_21408794 | 1.10 |
ENSDART00000140161
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr12_+_1139690 | 1.09 |
ENSDART00000160442
|
CABZ01072885.1
|
|
| chr3_+_29714775 | 1.08 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr17_-_29119362 | 1.05 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr10_+_45089820 | 1.03 |
ENSDART00000175481
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr25_-_13839743 | 1.01 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr16_+_46111849 | 1.01 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr1_+_33969015 | 1.00 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr18_+_1703984 | 1.00 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr7_-_28148310 | 0.99 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr17_+_15433518 | 0.97 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_-_31278048 | 0.95 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr17_+_15433671 | 0.95 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr9_+_24159280 | 0.95 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr10_+_22381802 | 0.95 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr6_-_51386656 | 0.93 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr13_-_29421331 | 0.91 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr6_-_11768198 | 0.91 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr11_-_1509773 | 0.90 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr25_+_7671640 | 0.90 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr20_+_40457599 | 0.88 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr2_-_9059955 | 0.88 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr8_-_39952727 | 0.86 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr16_-_28856112 | 0.84 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr12_-_26383242 | 0.83 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr2_+_20430366 | 0.82 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr20_+_52389858 | 0.81 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr24_+_24461341 | 0.81 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr2_-_30668580 | 0.80 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr10_-_2524297 | 0.80 |
ENSDART00000192475
|
CU856539.1
|
|
| chr6_+_7444899 | 0.79 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr3_-_30061985 | 0.79 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr23_+_16638639 | 0.79 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr1_+_12766351 | 0.79 |
ENSDART00000165785
|
pcdh10a
|
protocadherin 10a |
| chr5_-_21422390 | 0.78 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr17_+_13664442 | 0.78 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr8_+_31821396 | 0.77 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr1_+_6172786 | 0.76 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr15_+_22435460 | 0.76 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr18_-_38088099 | 0.74 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr14_-_4556896 | 0.73 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr19_-_30524952 | 0.73 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr9_-_51436377 | 0.72 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr11_+_30663300 | 0.72 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr15_-_15449929 | 0.72 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr5_-_67911111 | 0.72 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr20_+_32552912 | 0.71 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr3_-_48716422 | 0.70 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr8_+_10561922 | 0.70 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr19_+_5480327 | 0.69 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr24_-_7697274 | 0.69 |
ENSDART00000186077
|
syt5b
|
synaptotagmin Vb |
| chr2_+_29491314 | 0.68 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr10_-_34870667 | 0.67 |
ENSDART00000161272
|
dclk1a
|
doublecortin-like kinase 1a |
| chr16_-_44945224 | 0.66 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr8_-_34051548 | 0.66 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr7_+_19552381 | 0.66 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr9_+_29643036 | 0.65 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr9_+_24159725 | 0.64 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr10_-_34871737 | 0.64 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr24_+_24461558 | 0.64 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr6_+_49551614 | 0.63 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr20_+_19512727 | 0.61 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr8_+_26141680 | 0.61 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr10_-_2524917 | 0.60 |
ENSDART00000188642
|
CU856539.1
|
|
| chr19_-_5865766 | 0.59 |
ENSDART00000191007
|
LO018585.1
|
|
| chr14_+_3411771 | 0.59 |
ENSDART00000164778
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_-_41004731 | 0.58 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr23_-_6660985 | 0.58 |
ENSDART00000162405
|
CR450824.2
|
|
| chr22_-_13466246 | 0.58 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr20_+_32523576 | 0.58 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr1_-_19215336 | 0.58 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr24_+_15020402 | 0.57 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr2_+_2223837 | 0.57 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr16_+_33593116 | 0.57 |
ENSDART00000013148
|
pou3f1
|
POU class 3 homeobox 1 |
| chr2_+_38554260 | 0.57 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr3_-_56456527 | 0.56 |
ENSDART00000156553
|
cyth1a
|
cytohesin 1a |
| chr12_+_31735159 | 0.55 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr4_-_2219705 | 0.54 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr19_-_41371978 | 0.54 |
ENSDART00000166063
ENSDART00000170343 |
slc25a13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr23_+_30730121 | 0.53 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr11_+_41242644 | 0.52 |
ENSDART00000172008
|
pax7a
|
paired box 7a |
| chr22_-_22416337 | 0.52 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr7_-_71829649 | 0.52 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr17_-_42213285 | 0.51 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr14_-_17306261 | 0.51 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr3_+_34919810 | 0.51 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr9_-_37749973 | 0.50 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr1_+_6862917 | 0.50 |
ENSDART00000182953
|
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr7_+_13382852 | 0.50 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr10_-_43404027 | 0.50 |
ENSDART00000086227
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr13_-_29420885 | 0.50 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr5_+_21144269 | 0.49 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr5_-_34185115 | 0.49 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr19_-_20482261 | 0.49 |
ENSDART00000056205
|
satb1a
|
SATB homeobox 1a |
| chr16_-_29480335 | 0.49 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr4_-_75157223 | 0.49 |
ENSDART00000174127
|
CABZ01066312.1
|
|
| chr13_+_11440389 | 0.49 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr19_+_24872159 | 0.48 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr11_+_28476298 | 0.48 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr7_+_26629084 | 0.47 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr3_+_39853788 | 0.47 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr11_+_30057762 | 0.47 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr5_+_19712011 | 0.46 |
ENSDART00000131924
|
fam222a
|
family with sequence similarity 222, member A |
| chr22_-_26865361 | 0.46 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr23_+_4253957 | 0.46 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr15_-_18162647 | 0.45 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr1_-_21297748 | 0.45 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr6_+_51773873 | 0.45 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr13_+_18371208 | 0.45 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr25_-_25058508 | 0.44 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr21_-_17482465 | 0.44 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr6_+_40591149 | 0.44 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr5_+_44064764 | 0.43 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr24_+_20969358 | 0.43 |
ENSDART00000129675
|
drd3
|
dopamine receptor D3 |
| chr15_+_22867174 | 0.43 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr20_+_4060839 | 0.43 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr17_-_2386569 | 0.43 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr8_+_25959940 | 0.42 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr10_-_5844915 | 0.42 |
ENSDART00000185929
|
ankrd55
|
ankyrin repeat domain 55 |
| chr21_-_18648861 | 0.41 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr9_+_33009284 | 0.41 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr4_+_12111154 | 0.41 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr17_+_11675362 | 0.41 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr19_-_11106315 | 0.41 |
ENSDART00000059102
|
arhgef1a
|
Rho guanine nucleotide exchange factor (GEF) 1a |
| chr6_-_8377055 | 0.40 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr23_-_18707418 | 0.40 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr4_-_75158035 | 0.40 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr13_+_40034176 | 0.40 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr16_+_54209504 | 0.40 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr5_+_42400777 | 0.40 |
ENSDART00000183114
|
BX548073.8
|
|
| chr13_+_15656042 | 0.39 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr20_-_20930926 | 0.39 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr18_+_924949 | 0.39 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr7_+_10701770 | 0.39 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr21_+_13387965 | 0.39 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr20_-_20931197 | 0.39 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr3_-_47876427 | 0.38 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr11_+_7432533 | 0.38 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr13_+_33282095 | 0.38 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr15_+_23550890 | 0.38 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr9_+_2574122 | 0.38 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr9_+_36314867 | 0.37 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr15_+_32297441 | 0.37 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr25_-_6432463 | 0.37 |
ENSDART00000110389
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr1_-_15797663 | 0.37 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr11_-_25733910 | 0.37 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr5_+_42379517 | 0.35 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr1_+_38858399 | 0.35 |
ENSDART00000165454
|
CU915762.1
|
|
| chr22_-_26865181 | 0.35 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr5_+_42386705 | 0.35 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 0.35 |
ENSDART00000188489
|
BX548073.11
|
|
| chr15_-_46718759 | 0.35 |
ENSDART00000085926
ENSDART00000154577 |
zgc:162872
|
zgc:162872 |
| chr5_+_42393896 | 0.34 |
ENSDART00000189550
|
BX548073.13
|
|
| chr7_+_71547747 | 0.34 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr14_-_2933185 | 0.34 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_+_16564921 | 0.34 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr17_-_39772999 | 0.34 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr7_+_10701938 | 0.33 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr20_-_28642061 | 0.33 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr23_-_10048533 | 0.32 |
ENSDART00000166663
|
plxnb1a
|
plexin b1a |
| chr5_+_32009542 | 0.32 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
| chr21_+_27513859 | 0.32 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr17_-_40956035 | 0.32 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr4_-_20081621 | 0.32 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr13_+_31205439 | 0.31 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr11_+_40812590 | 0.31 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr12_-_41684729 | 0.30 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr16_+_28932038 | 0.30 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr21_-_275377 | 0.30 |
ENSDART00000157509
|
rln1
|
relaxin 1 |
| chr5_-_26566435 | 0.30 |
ENSDART00000146070
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr24_-_33873451 | 0.30 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr2_+_33368414 | 0.29 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr13_+_31211050 | 0.29 |
ENSDART00000111705
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr5_-_66397688 | 0.29 |
ENSDART00000161483
|
hip1rb
|
huntingtin interacting protein 1 related b |
| chr12_-_48006835 | 0.29 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr10_+_40214877 | 0.29 |
ENSDART00000109689
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr17_+_43623598 | 0.28 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr7_-_25895189 | 0.28 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr6_-_20952187 | 0.28 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr9_+_10014514 | 0.28 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr9_-_38021889 | 0.28 |
ENSDART00000183482
ENSDART00000124333 |
adcy5
|
adenylate cyclase 5 |
| chr22_-_31517300 | 0.28 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr23_-_30954738 | 0.28 |
ENSDART00000188996
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr9_+_10014817 | 0.27 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr14_+_6615564 | 0.27 |
ENSDART00000139292
|
si:dkeyp-44a8.2
|
si:dkeyp-44a8.2 |
| chr17_+_28533102 | 0.27 |
ENSDART00000156218
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr21_+_34976600 | 0.27 |
ENSDART00000191672
ENSDART00000139635 |
rbm11
|
RNA binding motif protein 11 |
| chr13_-_12006007 | 0.27 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr19_+_41464870 | 0.27 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx3+lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.4 | 1.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.3 | 0.8 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 1.6 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 0.7 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 0.5 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 0.8 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 1.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 0.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.8 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.1 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.5 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.3 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 1.0 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 1.1 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 2.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.5 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.7 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.5 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.3 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 0.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 2.0 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.5 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 3.1 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.4 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.3 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 1.0 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.3 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 1.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 1.3 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.3 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.7 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 2.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |