Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
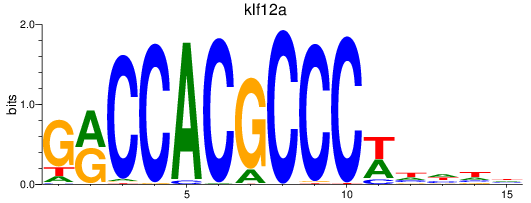
Results for klf12a
Z-value: 0.98
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
|
klf12a
|
ENSDARG00000115152 | Kruppel-like factor 12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12a | dr11_v1_chr1_+_34527213_34527213 | 0.06 | 9.3e-01 | Click! |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58221163 | 2.19 |
ENSDART00000157939
|
FO704813.1
|
|
| chr1_+_57371447 | 1.54 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr10_+_1668106 | 1.38 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr1_-_59176949 | 1.17 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr9_+_42095220 | 1.11 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr2_+_413370 | 1.03 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr17_-_4318393 | 0.95 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr10_+_19554604 | 0.84 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr5_+_37056818 | 0.83 |
ENSDART00000036760
|
tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr5_-_46273938 | 0.82 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr3_+_5575313 | 0.79 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr2_-_2020044 | 0.77 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr10_+_15777064 | 0.74 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_+_59015878 | 0.71 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr9_-_296169 | 0.71 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr15_+_24388782 | 0.70 |
ENSDART00000191661
ENSDART00000179995 ENSDART00000111226 |
sez6b
|
seizure related 6 homolog b |
| chr6_-_17849786 | 0.69 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr7_+_23907692 | 0.69 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr5_-_69482891 | 0.68 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr21_+_1382078 | 0.68 |
ENSDART00000188463
|
TCF4
|
transcription factor 4 |
| chr7_+_30867008 | 0.67 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr18_+_26895994 | 0.66 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr10_+_1638876 | 0.66 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr6_-_52156427 | 0.64 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr2_-_44282796 | 0.63 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr25_-_25736958 | 0.63 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr11_-_4235811 | 0.62 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr13_+_36764715 | 0.57 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr13_+_4405282 | 0.57 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr8_-_4618653 | 0.57 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr16_-_6821927 | 0.56 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr15_+_47161917 | 0.55 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr23_+_44732863 | 0.54 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr20_-_53366137 | 0.53 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr15_-_19250543 | 0.53 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr20_+_20672163 | 0.52 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr16_+_53455638 | 0.50 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr16_+_29303971 | 0.49 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr7_+_69528850 | 0.48 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr11_-_38083397 | 0.48 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr13_-_30028103 | 0.47 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr22_-_38516922 | 0.47 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr25_+_35375848 | 0.47 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr7_-_6592142 | 0.45 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr11_+_25064519 | 0.45 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr10_+_15777258 | 0.43 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr18_-_14677936 | 0.43 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr17_+_51764310 | 0.42 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr11_+_35364445 | 0.41 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr13_+_421231 | 0.41 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr10_+_37500234 | 0.40 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr2_+_30916188 | 0.40 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr11_-_42554290 | 0.40 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr3_-_16227490 | 0.39 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr15_-_19128705 | 0.38 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr21_+_34167178 | 0.38 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr2_+_23222939 | 0.38 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr11_+_1796426 | 0.37 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr23_-_17470146 | 0.37 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr10_+_39952995 | 0.37 |
ENSDART00000183077
|
BX927333.1
|
|
| chr16_-_45327616 | 0.36 |
ENSDART00000158733
|
si:dkey-33i11.1
|
si:dkey-33i11.1 |
| chr1_-_55058795 | 0.35 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr16_-_44399335 | 0.35 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr14_-_24391424 | 0.35 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr12_+_49125510 | 0.35 |
ENSDART00000185804
|
FO704607.1
|
|
| chr3_-_16227683 | 0.33 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr6_+_59967994 | 0.33 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr17_-_43466317 | 0.32 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr18_+_21122818 | 0.32 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr22_-_881080 | 0.32 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr9_-_48397702 | 0.32 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr4_-_22519516 | 0.31 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr8_-_410728 | 0.31 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr9_+_22375779 | 0.30 |
ENSDART00000183956
|
dgkg
|
diacylglycerol kinase, gamma |
| chr6_+_34512313 | 0.30 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr16_+_23282655 | 0.30 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr12_+_42436328 | 0.29 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr23_-_31645760 | 0.29 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_+_26019426 | 0.28 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr3_-_22829710 | 0.28 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr4_-_9557186 | 0.28 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr19_-_31686252 | 0.28 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr21_+_11415224 | 0.27 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr19_+_342094 | 0.27 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr7_-_18601206 | 0.27 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr17_+_11507131 | 0.27 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr13_+_36622100 | 0.27 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr1_-_50710468 | 0.26 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr9_+_34425736 | 0.26 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr3_-_6767440 | 0.26 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr5_+_66132394 | 0.26 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr25_+_25737386 | 0.26 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr1_+_11977426 | 0.26 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr12_+_47909026 | 0.25 |
ENSDART00000192472
|
tbata
|
thymus, brain and testes associated |
| chr25_-_11088839 | 0.25 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr8_-_1698155 | 0.25 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr6_+_34511886 | 0.25 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr23_-_41762797 | 0.25 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr7_+_38762043 | 0.25 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr4_-_5019113 | 0.24 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr24_+_41931585 | 0.24 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr10_-_22831611 | 0.24 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr19_+_48111285 | 0.23 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr3_-_40054615 | 0.23 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr7_-_12596727 | 0.22 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr5_+_43458304 | 0.22 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr25_-_1720736 | 0.22 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr5_-_54792239 | 0.22 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_+_35097024 | 0.22 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr20_-_43786515 | 0.22 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr22_-_21676364 | 0.22 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr17_-_34963575 | 0.22 |
ENSDART00000145664
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr15_+_47418565 | 0.21 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr9_+_4378153 | 0.21 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr19_+_8144556 | 0.21 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr4_-_5018705 | 0.21 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr1_+_45707219 | 0.21 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr23_-_41762956 | 0.20 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr5_+_38263240 | 0.20 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr6_-_15604417 | 0.20 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr6_-_15604157 | 0.20 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr18_-_3166726 | 0.20 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr4_-_5597802 | 0.19 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr4_+_3980247 | 0.19 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr3_+_59117136 | 0.19 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr22_-_20924747 | 0.19 |
ENSDART00000185845
ENSDART00000179672 |
ell
|
elongation factor RNA polymerase II |
| chr18_+_23249519 | 0.18 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr14_+_33264303 | 0.18 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr21_-_39670375 | 0.18 |
ENSDART00000151567
|
sgk494b
|
uncharacterized serine/threonine-protein kinase SgK494b |
| chr6_+_59854224 | 0.18 |
ENSDART00000083499
|
kdm6al
|
lysine (K)-specific demethylase 6A, like |
| chr7_-_64152087 | 0.18 |
ENSDART00000165234
|
CR354398.1
|
|
| chr7_-_69647988 | 0.17 |
ENSDART00000169943
|
LO018231.1
|
|
| chr20_-_54381034 | 0.17 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr2_-_39675829 | 0.17 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr8_+_1187928 | 0.17 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr24_-_36238054 | 0.16 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr1_+_51329069 | 0.16 |
ENSDART00000187729
ENSDART00000193246 ENSDART00000186753 ENSDART00000190206 |
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr19_-_30420602 | 0.16 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr19_-_34011340 | 0.16 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr5_+_4006837 | 0.16 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr23_-_32129569 | 0.16 |
ENSDART00000167761
ENSDART00000139569 |
zgc:92658
|
zgc:92658 |
| chr8_+_104114 | 0.16 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr3_-_47876427 | 0.16 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr1_+_45839927 | 0.16 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr12_+_6391243 | 0.16 |
ENSDART00000152765
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr18_+_507618 | 0.16 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr5_+_61657702 | 0.16 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr18_+_507835 | 0.16 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr21_+_21279159 | 0.16 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr1_+_42874410 | 0.16 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr20_+_572037 | 0.15 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr12_+_30046320 | 0.15 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr22_-_20924564 | 0.15 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr19_+_27479563 | 0.15 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr11_-_36341189 | 0.15 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr10_+_3299829 | 0.15 |
ENSDART00000183684
|
zgc:56235
|
zgc:56235 |
| chr11_-_36341028 | 0.15 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr23_-_27822920 | 0.15 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr13_+_28618086 | 0.14 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr18_-_14917296 | 0.14 |
ENSDART00000098662
|
panx2
|
pannexin 2 |
| chr2_+_37134281 | 0.14 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr16_+_30002605 | 0.14 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr2_-_32513538 | 0.14 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr7_-_19364813 | 0.14 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr25_-_31423493 | 0.14 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr21_+_10794914 | 0.14 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr16_+_43077909 | 0.14 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr10_+_6383270 | 0.14 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr3_+_22578369 | 0.14 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr6_+_16031189 | 0.13 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr22_-_29586608 | 0.13 |
ENSDART00000059869
|
adra2a
|
adrenoceptor alpha 2A |
| chr14_-_47849216 | 0.13 |
ENSDART00000192796
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr24_-_14712427 | 0.13 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr12_+_11080776 | 0.13 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr11_-_11301283 | 0.13 |
ENSDART00000113311
ENSDART00000180466 |
col9a1a
|
collagen, type IX, alpha 1a |
| chr10_+_4875262 | 0.13 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr23_-_43393589 | 0.13 |
ENSDART00000183592
|
zgc:174862
|
zgc:174862 |
| chr6_+_29693492 | 0.12 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr5_+_67812062 | 0.12 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr23_-_41821825 | 0.12 |
ENSDART00000184725
|
GMEB2
|
si:ch73-302a13.2 |
| chr16_-_20294236 | 0.12 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr10_-_42131408 | 0.12 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr24_+_33622769 | 0.12 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr17_-_44440832 | 0.12 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr22_+_34430310 | 0.12 |
ENSDART00000109860
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr14_+_12110020 | 0.11 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr5_-_12031174 | 0.11 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr17_-_25831569 | 0.11 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr17_-_18898115 | 0.11 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr8_-_17167819 | 0.11 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr23_+_30730121 | 0.11 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr11_+_5499661 | 0.11 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr9_-_1434484 | 0.10 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr1_-_7659870 | 0.10 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr10_+_41668483 | 0.10 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr5_+_61658282 | 0.10 |
ENSDART00000188878
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_72136548 | 0.10 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr20_-_36617313 | 0.10 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr23_-_10745288 | 0.10 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr22_-_38224315 | 0.10 |
ENSDART00000165430
ENSDART00000140968 |
hps3
|
Hermansky-Pudlak syndrome 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.7 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.2 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.5 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.5 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.5 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 1.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.1 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.5 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.7 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.1 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.0 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |