Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
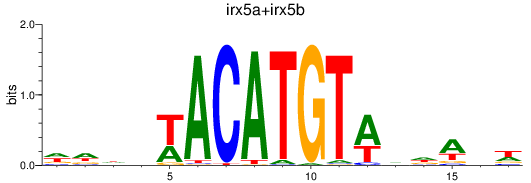
Results for irx5a+irx5b_irx3a+irx3b
Z-value: 1.55
Transcription factors associated with irx5a+irx5b_irx3a+irx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx5a
|
ENSDARG00000034043 | iroquois homeobox 5a |
|
irx5b
|
ENSDARG00000074070 | iroquois homeobox 5b |
|
irx3b
|
ENSDARG00000031138 | iroquois homeobox 3b |
|
irx3a
|
ENSDARG00000101076 | iroquois homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx3b | dr11_v1_chr25_-_36492779_36492779 | -0.94 | 1.7e-02 | Click! |
| irx5b | dr11_v1_chr25_+_36152215_36152215 | -0.73 | 1.6e-01 | Click! |
| irx3a | dr11_v1_chr7_+_36041509_36041509 | -0.28 | 6.5e-01 | Click! |
| irx5a | dr11_v1_chr7_-_35710263_35710263 | -0.22 | 7.2e-01 | Click! |
Activity profile of irx5a+irx5b_irx3a+irx3b motif
Sorted Z-values of irx5a+irx5b_irx3a+irx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50147948 | 1.12 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr11_-_45185792 | 0.86 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr3_-_32956808 | 0.71 |
ENSDART00000183902
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr1_-_7951002 | 0.70 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr7_+_56577522 | 0.62 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr7_+_56577906 | 0.59 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr17_-_5352924 | 0.56 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr20_+_34029820 | 0.55 |
ENSDART00000143901
ENSDART00000134305 |
prg4b
|
proteoglycan 4b |
| chr2_-_13333932 | 0.53 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr16_-_42186093 | 0.53 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr19_-_8748571 | 0.52 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr8_-_20914829 | 0.50 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr3_-_34100700 | 0.49 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr24_-_27419198 | 0.49 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr17_-_4395373 | 0.49 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr23_-_44574059 | 0.48 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr11_+_14284866 | 0.48 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr6_+_41144155 | 0.48 |
ENSDART00000191395
ENSDART00000143577 |
slc6a22.2
|
solute carrier family 6 member 22, tandem duplicate 2 |
| chr24_-_24983047 | 0.46 |
ENSDART00000066631
|
slc51a
|
solute carrier family 51, alpha subunit |
| chr16_+_42018367 | 0.46 |
ENSDART00000058613
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr3_-_16784280 | 0.45 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr8_+_3405612 | 0.45 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr18_-_49286381 | 0.45 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr8_-_4760723 | 0.45 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr2_+_17055069 | 0.45 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr10_-_42237304 | 0.45 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr3_-_33970190 | 0.44 |
ENSDART00000151238
|
ighj2-5
|
ighj2-5 |
| chr5_+_24087035 | 0.44 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr10_-_6976645 | 0.43 |
ENSDART00000123312
|
sh2d4a
|
SH2 domain containing 4A |
| chr7_+_34587081 | 0.43 |
ENSDART00000173817
|
fhod1
|
formin homology 2 domain containing 1 |
| chr6_-_7438584 | 0.43 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr9_-_5337923 | 0.43 |
ENSDART00000017939
|
tnfsf13b
|
TNF superfamily member 13b |
| chr11_+_13224281 | 0.43 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr19_-_48336535 | 0.42 |
ENSDART00000162752
|
si:ch73-359m17.6
|
si:ch73-359m17.6 |
| chr14_+_45471642 | 0.42 |
ENSDART00000126979
ENSDART00000172952 ENSDART00000173284 |
ubxn1
|
UBX domain protein 1 |
| chr22_+_6293563 | 0.42 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr22_-_1079773 | 0.41 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr11_+_24716837 | 0.40 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr3_-_32541033 | 0.40 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr22_-_26353916 | 0.39 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr13_+_2908764 | 0.39 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr1_+_53945934 | 0.39 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr12_+_20693743 | 0.39 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr24_-_23758003 | 0.39 |
ENSDART00000178085
|
BX640462.2
|
Danio rerio minichromosome maintenance domain containing 2 (mcmdc2), mRNA. |
| chr22_+_19478140 | 0.39 |
ENSDART00000135291
ENSDART00000136576 |
si:dkey-78l4.8
|
si:dkey-78l4.8 |
| chr25_-_3867990 | 0.38 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr13_+_13578552 | 0.38 |
ENSDART00000101673
|
foxi2
|
forkhead box I2 |
| chr6_-_54826061 | 0.37 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr3_-_33970431 | 0.37 |
ENSDART00000151313
|
ighj2-4
|
ighj2-4 |
| chr23_-_7594723 | 0.37 |
ENSDART00000115298
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr2_+_36608387 | 0.37 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr14_-_36799280 | 0.37 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr24_-_25098719 | 0.37 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr5_+_45677781 | 0.36 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr6_-_54180516 | 0.36 |
ENSDART00000149945
|
rps10
|
ribosomal protein S10 |
| chr25_-_29072162 | 0.36 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_-_58996324 | 0.36 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr7_+_9922607 | 0.36 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr25_-_34281411 | 0.36 |
ENSDART00000189932
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr19_-_7115229 | 0.36 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr18_+_22109379 | 0.36 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr7_-_71758307 | 0.35 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr16_-_53259409 | 0.35 |
ENSDART00000157080
|
si:ch211-269k10.4
|
si:ch211-269k10.4 |
| chr22_-_22340688 | 0.35 |
ENSDART00000105597
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr13_-_34683370 | 0.34 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr16_-_15263099 | 0.34 |
ENSDART00000125691
|
sntb1
|
syntrophin, basic 1 |
| chr17_+_22102791 | 0.34 |
ENSDART00000047772
|
mal
|
mal, T cell differentiation protein |
| chr16_-_47381519 | 0.34 |
ENSDART00000032188
ENSDART00000150136 |
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr19_-_11237125 | 0.34 |
ENSDART00000163921
|
ssr2
|
signal sequence receptor, beta |
| chr5_+_43782267 | 0.34 |
ENSDART00000130355
|
nos2a
|
nitric oxide synthase 2a, inducible |
| chr13_+_22717366 | 0.34 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr25_+_13205878 | 0.34 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr10_+_8629275 | 0.34 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr22_-_23666504 | 0.33 |
ENSDART00000158665
|
cfh
|
complement factor H |
| chr7_-_21928826 | 0.33 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr11_+_42474694 | 0.33 |
ENSDART00000056048
ENSDART00000184710 |
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr22_+_10781894 | 0.33 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr4_-_12997587 | 0.33 |
ENSDART00000140532
|
si:dkey-6a5.3
|
si:dkey-6a5.3 |
| chr17_+_30587333 | 0.33 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr16_+_29516098 | 0.32 |
ENSDART00000174895
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr3_-_50136424 | 0.32 |
ENSDART00000188843
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr15_-_21132480 | 0.31 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr13_-_41908583 | 0.31 |
ENSDART00000136515
|
ipmka
|
inositol polyphosphate multikinase a |
| chr7_-_6357952 | 0.31 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr8_-_36475328 | 0.31 |
ENSDART00000048448
|
si:busm1-266f07.2
|
si:busm1-266f07.2 |
| chr5_+_66170479 | 0.31 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr23_+_46157638 | 0.31 |
ENSDART00000076048
|
btr32
|
bloodthirsty-related gene family, member 32 |
| chr3_+_25999477 | 0.31 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr9_-_9415000 | 0.30 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr16_+_53259521 | 0.30 |
ENSDART00000155461
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr3_+_19336286 | 0.30 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr20_+_54299419 | 0.30 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr1_-_56032619 | 0.30 |
ENSDART00000143793
|
c3a.4
|
complement component c3a, duplicate 4 |
| chr4_+_7391400 | 0.30 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr16_-_16522013 | 0.30 |
ENSDART00000160602
|
nbeal2
|
neurobeachin-like 2 |
| chr14_+_21106444 | 0.30 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr20_+_10723292 | 0.30 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr15_-_563877 | 0.30 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr23_+_45027263 | 0.30 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr4_-_20313810 | 0.29 |
ENSDART00000136350
|
dcp1b
|
decapping mRNA 1B |
| chr22_+_37874691 | 0.29 |
ENSDART00000028565
|
ahsg1
|
alpha-2-HS-glycoprotein 1 |
| chr23_+_32044410 | 0.29 |
ENSDART00000048628
|
mylk2
|
myosin light chain kinase 2 |
| chr3_+_24618012 | 0.29 |
ENSDART00000111997
|
zgc:171506
|
zgc:171506 |
| chr9_+_24088062 | 0.29 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr14_+_15620640 | 0.29 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr20_-_33566640 | 0.29 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr24_-_27473771 | 0.29 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr3_+_21200763 | 0.29 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr20_+_24448007 | 0.29 |
ENSDART00000139866
|
si:dkey-273g18.1
|
si:dkey-273g18.1 |
| chr9_-_14055959 | 0.29 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr12_-_30549022 | 0.29 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr9_-_34396264 | 0.29 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr23_-_36313431 | 0.28 |
ENSDART00000125860
|
nfe2
|
nuclear factor, erythroid 2 |
| chr11_-_42134968 | 0.28 |
ENSDART00000187115
|
FP325130.1
|
|
| chr8_-_24252933 | 0.28 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr10_+_45345574 | 0.28 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr15_-_29573267 | 0.28 |
ENSDART00000099947
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr16_-_45230084 | 0.28 |
ENSDART00000184652
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr21_-_22648007 | 0.28 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr4_-_7869731 | 0.28 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr4_-_8043839 | 0.28 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr22_+_9069081 | 0.27 |
ENSDART00000187842
|
BX248395.1
|
|
| chr4_-_78020361 | 0.27 |
ENSDART00000159559
ENSDART00000162341 |
PGA3
|
pepsinogen 3, group I (pepsinogen A) |
| chr4_+_76755294 | 0.27 |
ENSDART00000142339
|
ms4a17a.12
|
membrane-spanning 4-domains, subfamily A, member 17A.12 |
| chr15_-_21155641 | 0.27 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr1_+_26667872 | 0.27 |
ENSDART00000152803
ENSDART00000152144 ENSDART00000152785 ENSDART00000152393 |
hemgn
|
hemogen |
| chr10_+_8656417 | 0.27 |
ENSDART00000123131
|
pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr1_-_23293261 | 0.27 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr1_-_55116453 | 0.27 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr4_+_62341346 | 0.27 |
ENSDART00000160601
|
znf1079
|
zinc finger protein 1079 |
| chr1_+_53919110 | 0.27 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr21_-_20832482 | 0.27 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr16_+_23984179 | 0.27 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr5_+_1933131 | 0.27 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr21_+_30084823 | 0.27 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr20_-_487783 | 0.27 |
ENSDART00000162218
|
col10a1b
|
collagen, type X, alpha 1b |
| chr15_+_12377887 | 0.26 |
ENSDART00000170769
|
il10ra
|
interleukin 10 receptor, alpha |
| chr8_-_1219815 | 0.26 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr21_-_41838284 | 0.26 |
ENSDART00000141067
|
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr9_+_29985010 | 0.26 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr9_+_426392 | 0.26 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr5_+_29831235 | 0.26 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr9_+_20853894 | 0.26 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr23_+_4890693 | 0.26 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr8_+_28695914 | 0.26 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr13_+_13681681 | 0.26 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr24_-_4765740 | 0.26 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr7_+_19600262 | 0.25 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr7_-_60831082 | 0.25 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr6_-_54180699 | 0.25 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr15_+_42560354 | 0.25 |
ENSDART00000059484
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr22_-_347424 | 0.25 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr16_-_44709832 | 0.25 |
ENSDART00000156784
|
si:ch211-151m7.6
|
si:ch211-151m7.6 |
| chr14_+_10567104 | 0.25 |
ENSDART00000160723
|
gpr174
|
G protein-coupled receptor 174 |
| chr4_+_18843015 | 0.25 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr12_-_31484677 | 0.24 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr14_+_21222287 | 0.24 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr23_+_19701587 | 0.24 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr4_-_17725008 | 0.24 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr1_+_1904419 | 0.24 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr16_+_25184207 | 0.24 |
ENSDART00000147584
|
hcst
|
hematopoietic cell signal transducer |
| chr10_+_40700311 | 0.24 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr14_+_1007169 | 0.24 |
ENSDART00000172676
|
f8
|
coagulation factor VIII, procoagulant component |
| chr20_+_54034512 | 0.24 |
ENSDART00000173226
|
si:dkey-241l7.2
|
si:dkey-241l7.2 |
| chr13_-_43149063 | 0.24 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr16_-_13388821 | 0.24 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr8_+_19356072 | 0.24 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr3_-_32925476 | 0.24 |
ENSDART00000189673
|
aoc2
|
amine oxidase, copper containing 2 |
| chr19_+_7115223 | 0.24 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr16_-_35975254 | 0.23 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr20_+_23440632 | 0.23 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr14_-_10617127 | 0.23 |
ENSDART00000154299
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr20_-_25518488 | 0.23 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr18_-_40773413 | 0.23 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr5_+_19261687 | 0.23 |
ENSDART00000139401
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr24_-_37640705 | 0.23 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr6_+_612594 | 0.23 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr14_+_21107032 | 0.23 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr3_-_50147160 | 0.23 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr2_-_127945 | 0.23 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr5_-_35159379 | 0.23 |
ENSDART00000144105
|
fcho2
|
FCH domain only 2 |
| chr17_+_24684778 | 0.23 |
ENSDART00000146309
ENSDART00000082237 |
znf593
|
zinc finger protein 593 |
| chr23_+_45845423 | 0.23 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr10_+_8401929 | 0.23 |
ENSDART00000059028
|
hvcn1
|
hydrogen voltage-gated channel 1 |
| chr21_-_34972872 | 0.22 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr2_-_42109575 | 0.22 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr6_-_15641686 | 0.22 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr20_+_54018005 | 0.22 |
ENSDART00000123026
ENSDART00000152894 |
si:dkey-241l7.5
|
si:dkey-241l7.5 |
| chr15_-_39971756 | 0.22 |
ENSDART00000063789
|
rps5
|
ribosomal protein S5 |
| chr9_+_21358941 | 0.22 |
ENSDART00000147619
ENSDART00000059402 |
eef1akmt1
|
EEF1A lysine methyltransferase 1 |
| chr24_-_25244637 | 0.22 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr1_+_40156908 | 0.22 |
ENSDART00000138886
|
si:ch211-113e8.9
|
si:ch211-113e8.9 |
| chr15_+_21202820 | 0.22 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr5_+_22370167 | 0.22 |
ENSDART00000145163
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr13_-_4223955 | 0.22 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr10_+_28428222 | 0.22 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr21_+_3796196 | 0.22 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr2_+_22496973 | 0.22 |
ENSDART00000159638
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr13_-_41901225 | 0.22 |
ENSDART00000167582
|
ipmka
|
inositol polyphosphate multikinase a |
| chr22_+_25672155 | 0.21 |
ENSDART00000087769
|
si:ch211-250e5.2
|
si:ch211-250e5.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx5a+irx5b_irx3a+irx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.4 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.4 | GO:0001782 | B cell homeostasis(GO:0001782) positive regulation of B cell proliferation(GO:0030890) |
| 0.1 | 0.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.4 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.7 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.1 | 0.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.4 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.3 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.1 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.3 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.2 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.2 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.3 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.4 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.3 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) cellular response to pH(GO:0071467) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) pore complex assembly(GO:0046931) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.3 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.6 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.4 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.1 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.2 | GO:1901185 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.8 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.3 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:0046348 | chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.3 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.0 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.2 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.4 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.1 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 0.6 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.2 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) excitatory extracellular ligand-gated ion channel activity(GO:0005231) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.0 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |