Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
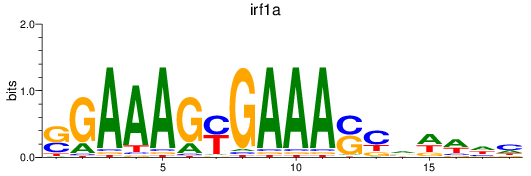
Results for irf1a
Z-value: 2.11
Transcription factors associated with irf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf1a
|
ENSDARG00000043492 | interferon regulatory factor 1a |
Activity profile of irf1a motif
Sorted Z-values of irf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7115229 | 3.84 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr5_+_13373593 | 3.75 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr13_+_13681681 | 2.88 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr19_+_7115223 | 2.83 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr11_-_20096018 | 2.50 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr1_-_52431220 | 2.03 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr15_-_29162193 | 1.97 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr1_-_52447364 | 1.95 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr23_+_642395 | 1.78 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr23_+_642001 | 1.73 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr14_+_36889893 | 1.70 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr1_-_8000428 | 1.68 |
ENSDART00000133098
|
si:dkey-79f11.5
|
si:dkey-79f11.5 |
| chr24_+_12945803 | 1.61 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr3_+_1942219 | 1.48 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr22_-_10580194 | 1.47 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr5_-_825920 | 1.45 |
ENSDART00000126982
|
zgc:158463
|
zgc:158463 |
| chr8_-_36469117 | 1.44 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr15_-_47929455 | 1.44 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr21_+_8533533 | 1.42 |
ENSDART00000077924
|
FO834888.1
|
|
| chr13_+_27328098 | 1.36 |
ENSDART00000037585
|
mb21d1
|
Mab-21 domain containing 1 |
| chr22_-_10158038 | 1.32 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr9_+_25330905 | 1.29 |
ENSDART00000101470
|
itm2bb
|
integral membrane protein 2Bb |
| chr21_-_22673758 | 1.29 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr19_+_7938121 | 1.26 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr6_+_52947699 | 1.26 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_29514473 | 1.26 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr3_+_29941777 | 1.24 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr20_+_26683933 | 1.21 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr4_+_5842433 | 1.20 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr5_-_57723929 | 1.20 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr22_-_36926342 | 1.16 |
ENSDART00000151804
|
si:dkey-37m8.11
|
si:dkey-37m8.11 |
| chr10_-_4375190 | 1.16 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr1_-_25177086 | 1.15 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr23_+_36063599 | 1.15 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr1_-_7930679 | 1.09 |
ENSDART00000146090
|
si:dkey-79f11.10
|
si:dkey-79f11.10 |
| chr19_+_23296616 | 1.08 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr2_-_42035250 | 1.05 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr4_-_12795030 | 1.05 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr17_+_35362851 | 1.03 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr1_-_47161996 | 1.03 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr19_+_7124337 | 1.03 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr19_-_325584 | 1.02 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr22_-_18164671 | 1.01 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr7_+_19600262 | 1.00 |
ENSDART00000007310
|
zgc:171731
|
zgc:171731 |
| chr6_+_52947186 | 0.96 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_1575435 | 0.96 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr22_-_18164835 | 0.95 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr3_-_48980319 | 0.89 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr22_-_17459587 | 0.88 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr13_+_47810211 | 0.84 |
ENSDART00000122964
|
zmp:0000001006
|
zmp:0000001006 |
| chr9_-_18215644 | 0.81 |
ENSDART00000145876
|
epsti1
|
epithelial stromal interaction 1 |
| chr22_-_15693085 | 0.81 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr23_+_46183410 | 0.80 |
ENSDART00000167596
ENSDART00000151149 ENSDART00000150896 |
btr31
|
bloodthirsty-related gene family, member 31 |
| chr4_-_17669881 | 0.80 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr16_-_51271962 | 0.79 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr17_-_35361322 | 0.79 |
ENSDART00000019617
|
rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr11_+_37638873 | 0.76 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr4_-_76370630 | 0.75 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr4_+_25558849 | 0.75 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr14_+_36414856 | 0.71 |
ENSDART00000123343
ENSDART00000015761 |
neil3
|
nei-like DNA glycosylase 3 |
| chr4_+_68776476 | 0.68 |
ENSDART00000169135
|
si:dkey-264f17.3
|
si:dkey-264f17.3 |
| chr7_-_60351876 | 0.67 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr11_-_19775182 | 0.66 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr5_-_41841675 | 0.66 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr23_+_5736226 | 0.65 |
ENSDART00000134527
ENSDART00000112220 |
ftr57
|
finTRIM family, member 57 |
| chr20_-_26936887 | 0.65 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr19_+_14115838 | 0.64 |
ENSDART00000192057
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr18_-_38270430 | 0.64 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr9_+_11293830 | 0.64 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr15_-_21702317 | 0.63 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr3_+_938438 | 0.62 |
ENSDART00000165671
|
irgf4
|
immunity-related GTPase family, f4 |
| chr22_+_29990448 | 0.62 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr2_+_42294944 | 0.61 |
ENSDART00000140599
ENSDART00000178811 |
ftr06
|
finTRIM family, member 6 |
| chr3_-_37148594 | 0.60 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr5_-_26247973 | 0.59 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr22_+_19230434 | 0.58 |
ENSDART00000132929
|
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr3_-_4760384 | 0.58 |
ENSDART00000108810
|
CABZ01046997.1
|
|
| chr13_-_50200042 | 0.58 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr23_-_6865946 | 0.57 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr3_-_34586403 | 0.57 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr18_-_38270077 | 0.57 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr7_+_54260004 | 0.56 |
ENSDART00000171909
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_-_23631400 | 0.56 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr25_+_8955530 | 0.55 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr3_-_10739625 | 0.55 |
ENSDART00000156144
|
zgc:112965
|
zgc:112965 |
| chr7_-_7984015 | 0.55 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr13_-_1151410 | 0.55 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr20_+_51199666 | 0.54 |
ENSDART00000169321
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr9_-_23765480 | 0.52 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr18_-_38270596 | 0.52 |
ENSDART00000098889
|
caprin1b
|
cell cycle associated protein 1b |
| chr22_+_36902352 | 0.52 |
ENSDART00000036273
|
trim35-20
|
tripartite motif containing 35-20 |
| chr19_+_4066449 | 0.52 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr4_+_70782916 | 0.52 |
ENSDART00000187960
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr5_-_26247671 | 0.52 |
ENSDART00000145187
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr11_+_1584747 | 0.50 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr20_-_20402500 | 0.48 |
ENSDART00000190747
ENSDART00000185065 |
prkchb
|
protein kinase C, eta, b |
| chr3_-_11040575 | 0.48 |
ENSDART00000159802
|
CR382337.2
|
|
| chr24_+_24809877 | 0.48 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr3_-_7997887 | 0.47 |
ENSDART00000172256
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr1_-_58002973 | 0.47 |
ENSDART00000140390
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr18_-_6534357 | 0.46 |
ENSDART00000192886
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr1_-_8966046 | 0.46 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr23_-_35649000 | 0.46 |
ENSDART00000053310
|
tmem18
|
transmembrane protein 18 |
| chr9_+_23765587 | 0.44 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr3_-_7974028 | 0.43 |
ENSDART00000144459
|
trim35-22
|
tripartite motif containing 35-22 |
| chr15_-_41749364 | 0.43 |
ENSDART00000155464
|
ftr73
|
finTRIM family, member 73 |
| chr5_-_38179220 | 0.42 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr14_-_48786708 | 0.42 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr5_+_38680061 | 0.42 |
ENSDART00000143259
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr13_-_50200348 | 0.41 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr3_+_8826905 | 0.41 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr10_-_45229877 | 0.41 |
ENSDART00000169281
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr3_-_3398383 | 0.40 |
ENSDART00000047865
|
si:dkey-46g23.2
|
si:dkey-46g23.2 |
| chr13_-_47810394 | 0.40 |
ENSDART00000105025
|
zmp:0000001138
|
zmp:0000001138 |
| chr5_+_38685089 | 0.40 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr4_+_53115165 | 0.39 |
ENSDART00000157470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr5_+_38679623 | 0.39 |
ENSDART00000131879
ENSDART00000170010 |
si:dkey-58f10.11
si:dkey-58f10.10
|
si:dkey-58f10.11 si:dkey-58f10.10 |
| chr12_-_13549538 | 0.38 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr4_-_25215968 | 0.38 |
ENSDART00000066932
ENSDART00000066933 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr18_+_50525109 | 0.35 |
ENSDART00000098390
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr18_-_6534516 | 0.34 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr16_-_43344859 | 0.34 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr8_-_12867128 | 0.34 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_-_47904043 | 0.32 |
ENSDART00000185328
ENSDART00000126740 |
ftr22
|
finTRIM family, member 22 |
| chr15_+_28355023 | 0.31 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr17_+_25833947 | 0.31 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr19_-_7706926 | 0.31 |
ENSDART00000151270
|
si:dkey-204a24.11
|
si:dkey-204a24.11 |
| chr4_+_70317135 | 0.31 |
ENSDART00000159286
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr3_-_7961226 | 0.30 |
ENSDART00000159801
|
trim35-23
|
tripartite motif containing 35-23 |
| chr1_+_58360935 | 0.30 |
ENSDART00000140754
|
si:dkey-222h21.1
|
si:dkey-222h21.1 |
| chr1_-_49530615 | 0.30 |
ENSDART00000113315
|
si:ch211-281g13.5
|
si:ch211-281g13.5 |
| chr19_+_12350069 | 0.30 |
ENSDART00000052240
|
psmg2
|
proteasome (prosome, macropain) assembly chaperone 2 |
| chr9_-_14084044 | 0.30 |
ENSDART00000141571
|
fer1l6
|
fer-1-like family member 6 |
| chr1_-_52201266 | 0.30 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr13_+_30912385 | 0.30 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr18_-_17724295 | 0.29 |
ENSDART00000121553
|
fam192a
|
family with sequence similarity 192, member A |
| chr12_-_4598237 | 0.28 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr2_-_43745146 | 0.28 |
ENSDART00000056164
ENSDART00000098052 |
ftr96
|
finTRIM family, member 96 |
| chr9_+_42269059 | 0.27 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr4_+_70967639 | 0.27 |
ENSDART00000159230
|
si:dkeyp-80d11.12
|
si:dkeyp-80d11.12 |
| chr7_+_26049818 | 0.27 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr4_-_50933168 | 0.27 |
ENSDART00000150296
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr24_-_8730913 | 0.27 |
ENSDART00000187228
ENSDART00000082349 ENSDART00000186660 |
tfap2a
|
transcription factor AP-2 alpha |
| chr16_-_50952266 | 0.26 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr4_+_279669 | 0.26 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr4_+_59841099 | 0.26 |
ENSDART00000144806
|
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr1_-_8141135 | 0.26 |
ENSDART00000152295
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr3_-_7940522 | 0.26 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr5_+_38726854 | 0.25 |
ENSDART00000138484
ENSDART00000142867 |
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr4_-_60790123 | 0.25 |
ENSDART00000137702
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr22_+_5858467 | 0.25 |
ENSDART00000137295
|
si:dkey-222f2.7
|
si:dkey-222f2.7 |
| chr2_+_42271830 | 0.25 |
ENSDART00000074027
ENSDART00000146444 |
ftr10
|
finTRIM family, member 10 |
| chr22_-_11520405 | 0.24 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr2_-_707152 | 0.24 |
ENSDART00000082304
|
ftr93
|
finTRIM family, member 93 |
| chr6_+_42338309 | 0.24 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr22_+_10158502 | 0.24 |
ENSDART00000005869
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr21_-_26715270 | 0.23 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr4_-_76079075 | 0.23 |
ENSDART00000158469
|
si:ch211-232d10.3
|
si:ch211-232d10.3 |
| chr3_+_1724941 | 0.23 |
ENSDART00000193402
|
BX321875.2
|
|
| chr1_+_50613868 | 0.23 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr22_-_6465060 | 0.23 |
ENSDART00000081687
|
zgc:172133
|
zgc:172133 |
| chr22_+_11520249 | 0.23 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr2_+_42236118 | 0.23 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr25_+_245018 | 0.23 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr4_+_33862438 | 0.23 |
ENSDART00000165044
|
si:dkey-28i19.3
|
si:dkey-28i19.3 |
| chr5_-_37820878 | 0.22 |
ENSDART00000165513
|
AL935065.1
|
Danio rerio uncharacterized LOC567472 (LOC567472), mRNA. |
| chr4_-_71177920 | 0.22 |
ENSDART00000158287
|
si:dkey-193i10.1
|
si:dkey-193i10.1 |
| chr17_-_28811747 | 0.22 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr18_-_12858016 | 0.22 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr4_-_13567387 | 0.22 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr3_+_36127287 | 0.22 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr2_+_42318012 | 0.21 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr4_+_70911498 | 0.21 |
ENSDART00000157674
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr1_-_52790724 | 0.21 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr17_-_23603263 | 0.21 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr3_+_48473346 | 0.21 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr6_-_19271210 | 0.20 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr4_+_71366326 | 0.20 |
ENSDART00000170807
|
si:ch211-76m11.8
|
si:ch211-76m11.8 |
| chr4_-_76027176 | 0.20 |
ENSDART00000165718
|
si:dkey-71l4.1
|
si:dkey-71l4.1 |
| chr10_+_2899108 | 0.20 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr5_-_26247215 | 0.19 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr18_+_14684115 | 0.19 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr20_-_36408836 | 0.19 |
ENSDART00000076419
|
lbr
|
lamin B receptor |
| chr6_+_56076595 | 0.18 |
ENSDART00000169747
|
rtf2
|
replication termination factor 2 domain containing 1 |
| chr18_-_14901437 | 0.18 |
ENSDART00000145842
ENSDART00000008035 |
trabd
|
TraB domain containing |
| chr2_-_38287987 | 0.18 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr5_-_23715027 | 0.17 |
ENSDART00000139020
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr18_+_14693682 | 0.17 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr23_+_39970425 | 0.17 |
ENSDART00000165376
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr2_+_49667767 | 0.16 |
ENSDART00000191013
ENSDART00000193757 ENSDART00000187134 ENSDART00000189100 |
BX323861.3
|
|
| chr10_+_40623414 | 0.16 |
ENSDART00000187616
|
zgc:172131
|
zgc:172131 |
| chr3_+_19336286 | 0.16 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr22_+_9994833 | 0.16 |
ENSDART00000172696
ENSDART00000192101 |
BX324216.1
|
|
| chr4_+_8016457 | 0.16 |
ENSDART00000014036
|
optn
|
optineurin |
| chr18_+_8812549 | 0.15 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr8_-_12867434 | 0.15 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr21_-_11632403 | 0.15 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr6_+_56076848 | 0.15 |
ENSDART00000059438
ENSDART00000157040 |
rtf2
|
replication termination factor 2 domain containing 1 |
| chr11_-_30341431 | 0.15 |
ENSDART00000078378
|
cflara
|
CASP8 and FADD-like apoptosis regulator a |
| chr7_+_22792895 | 0.14 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 1.3 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 9.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 0.8 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 1.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 1.0 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.5 | GO:0071635 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.6 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.1 | 2.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.6 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 1.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 3.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.5 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.6 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.2 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.3 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 3.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.7 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.8 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0051131 | proteasome assembly(GO:0043248) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.7 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.1 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 9.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 2.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 1.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.4 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 3.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 3.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 3.0 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |